Abstract
Squamous cell carcinoma is the second most common cutaneous malignancy. The diagnosis can occasionally be difficult as there are many lesions that are mimics, clinically and on pathologic examination. One of the most challenging lesions to differentiate from squamous cell carcinoma is pseudoepitheliomatous hyperplasia, a reactive proliferation of the epidermis that can be encountered secondary to a variety of inflammatory and neoplastic conditions. Utilizing the data set from our previously performed DNA microarray studies on formalin-fixed and paraffin-embedded tissue, we found that the genes C15orf48 and KRT9 had a distinct and robust gene expression pattern in distinguishing squamous cell carcinoma from pseudoepitheliomatous hyperplasia. C15orf48 had higher expression than KRT9 in squamous cell carcinoma, but lower expression than KRT9 in pseudoepitheliomatous hyperplasia. We developed and blindly validated a multiplex TaqMan PCR assay that utilizes these two highly discriminatory genes, which can be performed on material extracted from formalin-fixed and paraffin-embedded tissue. The TaqMan assay was able to differentiate squamous cell carcinoma from pseudoepitheliomatous hyperplasia in 54 of 58 cases (93%). Squamous cell carcinoma was accurately identified in 27 of 28 cases (96%); pseudoepitheliomatous hyperplasia in 27 of 30 cases (90%). This multiplex TaqMan PCR assay may be used as a helpful ancillary molecular diagnostic test to accurately distinguish squamous cell carcinoma from pseudoepitheliomatous hyperplasia in challenging cases.
Similar content being viewed by others
Main
Cutaneous squamous cell carcinoma is the second most common non-melanoma skin cancer with over 250 000 cases diagnosed per year.1 Owing to the potential to recur if incompletely excised and occasionally metastasize, an accurate diagnosis of squamous cell carcinoma is paramount. These concerns are magnified in cosmetically sensitive and/or high-risk locations, and in immunocompromised patients. There are a variety of preneoplastic and reactive clinicopathologic entities that resemble cutaneous squamous cell carcinoma that have been confounding both dermatologists and pathologists for decades.2 One of the most difficult of these lesions to differentiate from well-differentiated squamous cell carcinoma is pseudoepitheliomatous hyperplasia.
Pseudoepitheliomatous hyperplasia is a reactive proliferation of the epidermis that can be encountered secondary to a variety of inflammatory and neoplastic conditions.3, 4 Histologically, pseudoepitheliomatous hyperplasia is characterized by irregular strands, cords, and nests of squamoid cells extending into the dermis. These cells can exhibit substantial nuclear atypia and mitoses. The differentiation between a well-differentiated squamous cell carcinoma and pseudoepitheliomatous hyperplasia can be particularly difficult on superficial or poorly oriented biopsies. Occasional cases defy definitive classification even after exhaustive examination of multiple histologic sections and correlation with detailed clinical information. This can lead to additional biopsies or excisions that may not be necessary. To date, there are no highly discriminatory ancillary immunohistochemical or molecular tests to differentiate squamous cell carcinoma from pseudoepitheliomatous hyperplasia.
In our prior study,5 we utilized DNA microarrays and showed that squamous cell carcinoma and pseudoepitheliomatous hyperplasia had distinct molecular signatures. We examined some of the most significantly differentially expressed genes and molecular pathways that were enriched in comparing squamous cell carcinoma to pseudoepitheliomatous hyperplasia. From this data we developed a multiplex TaqMan PCR assay utilizing KRT9 and C15orf48 that differentiated squamous cell carcinoma from pseudoepitheliomatous hyperplasia in 54 of 58 cases (93%).
Materials and methods
Sample Selection
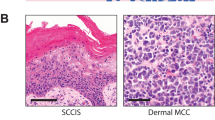
Thirty-five cases of pseudoepitheliomatous hyperplasia, inflammatory type, and thirty-three cases of cutaneous squamous cell carcinoma were identified from the Tamtron database. The slides and formalin-fixed paraffin-embedded tissue (<2 years old) were retrieved. The slides were reviewed by two of the authors (SR, SB) and their diagnoses confirmed. The squamous cell carcinomas were well-to-moderately differentiated (Figure 1) and arose from a background of sun damaged skin represented by associated actinic keratosis and/or solar elastosis. The cases of pseudoepitheliomatous hyperplasia were of the inflammatory type and represented by cases of prurigo nodularis and lichen simplex chronicus (Figure 2). The areas of interest were removed from the paraffin blocks with a sterile surgical scalpel.
RNA Isolation and Quality Control
Total RNA was isolated using the Ambion RecoverAll (Applied Biosystems/Ambion, Austin, TX, USA) kit according to the manufacturer’s instructions. Briefly, formalin-fixed and paraffin-embedded samples were deparaffinized using a series of xylene and ethanol washes, and then subjected to a proteinase K digestion at 50 °C for 16 h to release RNA from covalently linked proteins. Finally, total RNA was purified by capture on a glass-fiber filter. After washing, the total RNA was eluted. RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and purity/concentration was determined using a NanoDropTM 8000 (NanoDrop Products, Wilmington, DE, USA). The RNA samples with RNA integrity number ≥5 and 260:280 ratio ≥1.7 were selected for the analysis.
Multiplex TaqMan Real Time PCR Analysis
Two sets of primers and probes were designed targeting C15orf48 and KRT9 using Primer3plus online software as follows:
KRT9
Probe (FAM): 5′-GGCATATGTGCTCTGGCTTT-3′
Forward primer: 5′-GCCTGCTTATTGGATCCTGA-3′
Reverse primer: 5′-CAGGCCAGAGAGAGGAAAGA-3′
C15orf48
Probe (VIC): 5′-AAACCGATGTGATCCTT-3′
Forward primer: 5′-TGGAGCCTCATCTTTCGCTGTG-3′
Reverse primer: 5′-TGGGTTTCCATTGTTGGTTGATTG-3′.
Total RNA was first reverse transcribed into cDNA with Invitrogen SuperScript VILO cDNA Synthesis Kit (cat. no: 11754-050). Taqman qRT-PCR was then performed in a 20-μl volume with Invitrogen EXPRESS qPCR Supermix Universal kits (cat. no: 11785-200). To optimize the multiplex qRT-PCR performance, the primer-limiting tests were performed to find the optimal primer and probe concentration. We used a final concentration of 300 nM of primers and probes each in the reaction mix. The qRT-PCR was run with one cycle at 50 °C for 2 min (UDG incubation), one cycle at 95 °C for 2 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Two replicates were run for each sample. The analysis was performed by directly comparing CT values (cycle threshold, ie, the cycle where the increase in fluorescence crosses the threshold to be exponential) of two genes in the same reaction. All experiments were performed on an Applied Biosystem 7500 Fast Real-time PCR System.
Results
From the differentially expressed genes identified by our previous microarray analysis,5 we found that C15orf48 and KRT9 had a distinct and robust gene expression pattern in distinguishing squamous cell carcinoma from pseudoepitheliomatous hyperplasia. C15orf48 had higher expression than KRT9 in squamous cell carcinoma, but lower expression than KRT9 in pseudoepitheliomatous hyperplasia. We confirmed this finding with qRT-PCR utilizing the same RNA samples in the microarray experiment (data not shown) and then blindly tested five additional cases each of squamous cell carcinoma and pseudoepitheliomatous hyperplasia. Data consistently showed that KRT9 had a higher CT value than C15orf48 in squamous cell carcinoma, but lower CT value than C15orf48 in pseudoepitheliomatous hyperplasia (Figure 3). This inverse gene expression relationship offered unique diagnostic potential without data normalization through a TaqMan PCR assay utilizing these two genes. After validating this new assay, we tested 58 cases with two of the authors who performed the Taqman assay (XL and JZ) blinded to the final histologic diagnosis. The assay was able to differentiate squamous cell carcinoma from pseudoepitheliomatous hyperplasia in 54 of 58 cases (93%). Squamous cell carcinoma was accurately identified in 27 of 28 cases (96%) and pseudoepitheliomatous hyperplasia in 27 of 30 cases (90%) (Table 1).
Analysis of qRT-PCR results by cycle threshold value (CT, ie, the cycle where the increase in fluorescence crosses the threshold to be exponential). The raw qRT-PCR CT ratio between KRT9 and C15orf48 was calculated. Squamous cell carcinoma samples had a positive CT ratio, whereas pseudoepitheliomatous hyperplasia samples had a negative CT ratio indicating that C15orf48 had higher expression level than KRT9 in squamous cell carcinoma, but lower expression level than KRT9 in pseudoepitheliomatous hyperplasia.
Discussion
Pseudoepitheliomatous hyperplasia is a known clinicopathologic simulator of cutaneous squamous cell carcinoma. Accurate diagnosis is essential to guide clinical management. Our prior study5 demonstrated that squamous cell carcinoma and pseudoepitheliomatous hyperplasia are molecularly distinct lesions with unique gene signatures. However, as the use of DNA microarrays to routinely distinguish between them is neither currently practical nor cost effective, we developed and blindly validated a TaqMan PCR assay utilizing two highly discriminatory genes (C15orf48 and KRT9). These two genes not only were significantly differentially expressed, but also have an inverse gene expression pattern in differentiating squamous cell carcinoma from pseudoepitheliomatous hyperplasia. The latter feature eliminates data normalization and is particularly important for clinical practice as one can interpret the data by simply looking at pictures without any need for calculations.
The TaqMan PCR assay was able to distinguish between squamous cell carcinoma and pseudoepitheliomatous hyperplasia in 54 of 58 cases (93%) and may be one of the most successful discriminatory ancillary tests developed to date. TaqMan PCR is a practical ancillary study because of its rapidity, cost-effectiveness, and capability of high throughput. It is already widely used in microbiology to identify bacterial pathogens and quantitate viral loads in HIV and hepatitis C. TaqMan PCR is also used in human leukocyte antigen typing.
There have been prior attempts to utilize immunohistochemistry to help distinguish between squamous cell carcinoma and pseudoepitheliomatous hyperplasia. One study used a panel of p53, E-cadherin, and matrix metalloproteinase-1 that helped to distinguish squamous cell carcinoma from pseudoepitheliomatous hyperplasia in head and neck biopsy specimens.6 However, another study demonstrated that p53 immunohistochemical expression in this context may be more of an indicator of immaturity and proliferative capacity of the cell rather than one of neoplasia or malignancy.7 None of these ancillary immunohistochemical stains are widely used in everyday clinical practice. Additional immunohistochemical studies evaluating the number of Langerhans cells with CD1a and utilizing the differential expression of proliferating cell nuclear antigen were not successful in distinguishing squamous cell carcinoma from pseudoepitheliomatous hyperplasia.8, 9
There were 4 out of 58 cases where the PCR test gave conflicting results. Squamous cell carcinoma was not accurately identified in 1 of 28 cases (4%) and pseudoepitheliomatous hyperplasia in 3 of 30 cases (10%). These false negative squamous cell carcinoma and false positive pseudoepitheliomatous hyperplasia cases were re-reviewed and their histologic diagnoses confirmed in light of the PCR test results. These four cases did not exhibit any different or unusual histopathologic features. In addition, by comparing the paraffin blocks to the corresponding glass slides, we confirmed that the appropriate areas from the blocks had been sampled for RNA isolation. Although our PCR test was highly successful, we believe the discrepant cases reveal some limitations of our study.
The cases of squamous cell carcinoma selected for the current study were well-to-moderately differentiated squamous cell carcinomas within a background of actinic damage. The squamous cell carcinomas examined by DNA microarray analysis were molecularly similar as previously demonstrated on hierarchical cluster analysis.5 As demonstrated in our previous study, numerous molecular pathways are deregulated in squamous cell carcinoma. The testing of a small sample of squamous cell carcinoma may not reveal all of the different pathways deregulated in squamous cell carcinoma. A common pathway is purported for squamous cell carcinomas arising from ultraviolet radiation.1 However, it is possible that other converging or alternate pathways because of additional contributing factors may lead to the development of squamous cell carcinoma. It is well known that there are many conditions other than ultraviolet radiation associated with the development of squamous cell carcinoma, including chronically injured or diseased skin, immunosuppression, chemical carcinogens, genodermatoses, and chronic inflammatory disorders.1 These other contributing factors may lead to carcinogenesis utilizing other pathways.
The cases of pseudoepitheliomatous hyperplasia selected for the current study were inflammatory types including lichen simplex chronicus and prurigo nodularis. The cases of pseudoepitheliomatous hyperplasia examined by DNA microarray analysis were molecularly similar as previously demonstrated on hierarchical cluster analysis.5 As described with squamous cell carcinoma, the testing of a small sample may not accurately reflect the myriad of different pathways deregulated in pseudoepitheliomatous hyperplasia. Pseudoepitheliomatous hyperplasia is a heterogeneous group of entities for which there are likely differing molecular pathways. It is well known that there are many different conditions associated with the development of pseudoepitheliomatous hyperplasia, including infections, inflammatory conditions, and benign and malignant neoplasms. This heterogeneity may account for the higher number of cases that were incorrectly classified as squamous cell carcinoma.
The distinct and robust gene expression pattern of C15orf48 and KRT9 in distinguishing squamous cell carcinoma from pseudoepitheliomatous hyperplasia raises questions about the role of these genes in the pathogenesis of these lesions. From our previous DNA microarray studies, we found that KRT9 was one of the most significantly downregulated genes in comparing squamous cell carcinoma to pseudoepitheliomatous hyperplasia (fold change=−15.30).5 Keratinocytes have an extensive intracellular keratin filament network. Loss of regulation of this network may be necessary for the evolution of squamous cell carcinoma from normal skin.5 KRT9 encodes an intermediate filament chain expressed only in the terminally differentiated epidermis of palms and soles. Mutations in this gene cause epidermolytic palmoplantar keratoderma.10 Its role in tumorigenesis is currently unknown and has not been previously described in association with squamous cell carcinoma. Our previous DNA microarray studies also showed C15orf48 (NMES1) to be significantly upregulated in comparing squamous cell carcinoma to pseudoepitheliomatous hyperplasia (fold change=10.99).5 C15orf48 is expressed in the esophagus, stomach, small intestine, colon, and placenta of adult human tissues. The function of C15orf48 is unknown, although it may have a role in the pathogenesis of squamous cell carcinoma. Downregulation of C15orf48 has been demonstrated in esophageal squamous cell carcinomas11 and its aberrant methylation reported in cervical squamous cell carcinoma cell lines.12
In summary, we have developed a simple, reliable, and low-cost multiplex TaqMan PCR assay that was able to distinguish squamous cell carcinoma from pseudoepitheliomatous hyperplasia in the vast majority of cases examined. Our study demonstrates the practical use of an ancillary study for assistance in distinguishing cases of squamous cell carcinoma from pseudoepitheliomatous hyperplasia in a timely and cost-effective manner. This study also provides a framework for future research. We realize that our current model is restrictive in examining only actinically damaged squamous cell carcinoma and inflammatory types of pseudoepitheliomatous hyperplasia. We plan on analyzing different types of squamous cell carcinoma and pseudoepitheliomatous hyperplasia on DNA microarrays in order to further refine the gene signatures that are differentially expressed between squamous cell carcinoma and pseudoepitheliomatous hyperplasia, the different types of squamous cell carcinoma and the different types of pseudoepitheliomatous hyperplasia. Also, more TaqMan PCR studies are being performed on cases of squamous cell carcinoma and pseudoepitheliomatous hyperplasia with expansion to the different subtypes of squamous cell carcinoma and pseudoepitheliomatous hyperplasia as previously outlined. Also, studies are needed on cases that are histologically ambiguous with long-term clinical follow-up to accurately resolve the diagnostic challenges between squamous cell carcinoma and pseudoepitheliomatous hyperplasia.
References
Alam M, Ratner D . Cutaneous squamous-cell carcinoma. N Eng J Med 2001;344:975–983.
Grunwald MH, Lee JY, Ackerman AB . Pseudocarcinomatous hyperplasia. Am J Dermatopathol 1988;10:95–103.
El-Khoury J, Kibbi AG, Abbas O . Mucocutaneous pseudoepitheliomatous hyperplasia: a review. Am J Dermatopathol 2012;34:165–175.
Zayour M, Lazova R . Pseudoepitheliomatous hyperplasia: a review. Am J Dermatopathol 2011;33:112–122.
Ra SH, Su A, Li X et al. Molecularly enriched pathways and differentially expressed genes distinguishing cutaneous squamous cell carcinoma from pseudoepitheliomatous hyperplasia. Diagn Mol Pathol 2013;22:41–47.
Zarovnaya E, Black C . Distinguishing pseudoepitheliomatous hyperplasia from squamous cell carcinoma in mucosal biopsy specimens from the head and neck. Arch Pathol Lab Med 2005;129:1032–1036.
Lee YS, Teh M . p53 expression in pseudoepitheliomatous hyperplasia, keratoacanthoma, and squamous cell carcinoma of skin. Cancer 1994;73:2317–2323.
Galan A, Ko CJ . Langerhans cells in squamous cell carcinoma vs. pseudoepitheliomatous hyperplasia of the skin. J Cutan Pathol 2007;34:950–952.
Li J, Lee YS . Proliferating cell nuclear antigen (PCNA) expression in pseudoepitheliomatous hyperplasia, keratoacanthoma and squamous cell carcinoma of the skin. Ann Acad Med Singapore 1996;25:526–530.
Reis A, Hennies HC, Langbein L et al. Keratin 9 gene mutations in epidermolytic palmoplantar keratoderma (EPPK). Nat Genet 1994;6:174–179.
Zhou J, Wang H, Lu A et al. A novel gene, NMES1, downregulated in human esophageal squamous cell carcinoma. Int J Cancer 2002;101:311–316.
Sova P, Feng Q, Geiss G et al. Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol Biomarkers Prev 2006;15:114–123.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
SB, SR, and XL have a patent through UCLA on the above-mentioned TaqMan PCR assay.
Rights and permissions
About this article
Cite this article
Su, A., Ra, S., Li, X. et al. Differentiating cutaneous squamous cell carcinoma and pseudoepitheliomatous hyperplasia by multiplex qRT-PCR. Mod Pathol 26, 1433–1437 (2013). https://doi.org/10.1038/modpathol.2013.82
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.82