Abstract
Proliferation rates in diffuse large B-cell lymphoma have been associated with conflicting outcomes in the literature, more often with high proliferation associated with poor prognosis. In most studies, the proliferation rate was estimated by a pathologist using an immunohistochemical stain for the monoclonal antibody Ki-67. We hypothesized that a quantitative image analysis algorithm would give a more accurate estimate of the proliferation rate, leading to better associations with survival. In all, 84 cases of diffuse large B-cell lymphoma were selected according to the World Health Organization criteria. Ki-67 percentage positivity estimated by the pathologist was recorded from the original report. The same slides were then scanned using an Aperio ImageScope, and Ki-67 percentage positivity was calculated using a computer-based quantitative immunohistochemistry nuclear algorithm. In addition, chart review was performed and survival time was recorded. The Ki-67 percentage estimated by the pathologist from the original report versus quantitative image analysis was significantly correlated (P<0.001), but pathologist Ki-67 percentages were significantly higher than quantitative image analysis (P=0.021). There was less agreement at lower Ki-67 percentages. Comparison of Ki-67 percentage positivity versus survival did not show significant association either with pathologist estimate or quantitative image analysis. However, although not significant, there was a trend of worse survival at higher proliferation rates detected by the pathologist but not by quantitative image analysis. Interestingly, our data suggest that the Ki-67 percentage positivity as assessed by the pathologist may be more closely associated with survival outcome than that identified by quantitative image analysis. This may indicate that pathologists are better at selecting appropriate areas of the slide. More cases are needed to assess whether this finding would be statistically significant. Due to the good correlation between pathologist estimate and quantitative image analysis, there is no substantial benefit to using quantitative image analysis at this point of time.
Similar content being viewed by others
Main
Diffuse large B-cell lymphoma is the most frequent type of non-Hodgkin lymphoma in adults in the United States.1 Due to its heterogeneity in appearance and clinical behavior, numerous studies have attempted to further subclassify this entity into meaningfully distinct groups. Although gene expression profiling and surrogate immunohistochemical studies have elicited significant differences in pathogenesis and overall survival,2, 3, 4, 5, 6, 7, 8, 9 with groups such as those of germinal center B-cell origin and activated B-cell types identified, the majority of these studies took place before the routine addition of rituximab to the chemotherapeutic regimen. The relatively recent use of chemotherapy regimens incorporating rituximab (R-CHOP) has shown in some studies an eradication of these differences in prognostic markers,10 although some recent studies have suggested no significant difference between CHOP and R-CHOP groups.11 Although many practicing pathologists have avoided the routine classification of diffuse large B-cell lymphoma into germinal center B-cell and activated B-cell phenotypes, for reasons including those mentioned above, most pathologists still continue to report the proliferation rate of the tumor. The Ki-67 monoclonal antibody is commonly used to measure the proliferation index, and a higher proliferation rate is commonly associated with worse prognosis. Many of the studies, however, that report an association between high proliferation rate and poor prognosis12, 13, 14 were performed before the routine use of rituximab in chemotherapy regimens. Examination of the medical literature shows some conflicting data with some studies reporting no effect of proliferation rates on prognosis,15, 16 and other studies even showing correlation between low proliferation index and poor outcome.17 Despite the variable data, the most recent World Health Organization criteria on diffuse large B-cell lymphoma does list high proliferation index as a likely indicator of poorer prognosis.1
Traditionally in the clinical setting, pathologists have estimated this proliferation rate using a Ki-67 immunohistochemical stain, selecting areas of tumor involvement and estimating the percentage of tumor cells with positive nuclei. The presence of nuclear staining with Ki-67 indicates an actively cycling cell while quiescent cells should be negative.18 Although some research studies have used more complex and time-consuming methods involving counting cells in grids, in practical application one would generally study multiple areas of involvement and give a rough estimate without actually counting all of the cells. There is some debate as to which method is best and whether a semi-quantitative estimation is sufficient. No specific cutoff has been established at which prognosis changes, but several studies have used cutoffs in the range of 60–80% Ki-67 positivity to achieve statistical significance.12, 14, 19
In recent years, virtual microscopy and quantitative image analysis have become viable options in the clinical laboratory and have been used both in clinical and research applications, including areas of high-volume study such as routine gynecological cytology, where their cost savings is more obvious.20, 21, 22, 23, 24 In areas of pathology such as the microscopical evaluation of lymph nodes for malignancy, quantitative image analysis is not widely used and less literature exists as to its potential benefits. In the study of large cell lymphomas, proliferation indices are frequently evaluated to assess for potentially aggressive tumors, although as stated above, it is typical for only a semiquantitative estimate to be given in clinical practice.
In this study, we sought to address whether the use of computer-driven quantitative image analysis would add clinical value. We hypothesized that a quantitative image analysis system may be able to provide a more accurate proliferation index than that obtained by routine pathologist estimation. As previous reports in the literature show conflicting conclusions as to the prognostic significance of the proliferation rate, we further hypothesized that quantitative image analysis of the proliferation index would reveal a stronger correlation with prognosis, with higher proliferation conferring a worse prognosis. Lastly, we suspected that the number of background T-cells might confound the computer's ability to assess the Ki-67 percentage and to identify which cells were tumor and which were background T-cells. Thus, we also became interested in quantifying the number of background T-cells, as we thought an algorithm might be necessary to ‘subtract’ the non-tumor T-cells from the computer-generated Ki-67 proliferation index, thereby providing the ‘true’ proliferation index of the tumor. The ultimate question that this study addresses is whether the current pathologist assessment of proliferation rate is adequate or whether a computer-driven quantitative analysis would add prognostic value, and should be implemented in routine clinical laboratory evaluation of diffuse large B-cell lymphoma.
Materials and methods
Case Selection
This study was approved by the Human Research Protections Office of the University of New Mexico Health Sciences Center. Cases were selected from the pathology database that met the 2008 World Health Organization criteria for the diagnosis of diffuse large B-cell lymphoma, not otherwise specified. Cases of central nervous system lymphoma, post-transplant lymphoproliferative disorders and cutaneous lymphomas were excluded. Cases classified as T-cell/histiocyte-rich large B-cell lymphoma were also excluded from the study. None of the cases were diagnostic of Burkitt lymphoma, nor did any of the cases evaluated by cytogenetics have the ‘double hit’ finding of both a MYC rearrangement and a t(14;18) translocation, which has been associated with more aggressive biological behavior and may ultimately be classified as a separate entity.1, 25 We specifically selected cases no earlier than 2001 to better standardize the treatment that patients received. In all cases, treatment was potentially curative and where indicated, the R-CHOP regimen (rituxan, cyclophosphamide, doxorubicin, vincristine and prednisone) was used. After exclusion criteria were used, a total of 84 cases were collected. Of these, 50 cases originated in a lymph node while 34 cases were extranodal.
Immunohistochemistry
Archived glass slides from each case were collected. Standard laboratory protocols were used in the staining of each case as follows: Sections were cut at 4 to 5 μm intervals. One slide each was stained with hematoxylin & eosin, and a section from each case was also stained using antibodies to the following: Ki-67 (clone 30-9, pre-diluted, antigen retrieval CC1-30, Cell Marque, Rocklin, CA, USA) and CD3 (clone 2GV6, pre-diluted, antigen retrieval CC1-30, Ventana, Tucson, AZ, USA). The immunohistochemical stains were performed using standard protocols for the particular antibodies.
Digital Imaging and Quantitative Image Analysis
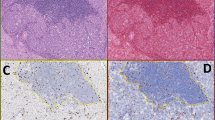
An H&E, CD3 and Ki-67 slides were scanned for each case using the Aperio ScanScope (Aperio Technologies, Vista, CA, USA). These were the same slides used to make the original diagnosis. After saving each digital image, all intact areas of tumor (excluding areas of significant crush artifact, necrosis and other poor quality regions) were selected for analysis. (Figure 1) Aperio ImageScope software (Aperio Technologies) was used, including both the Aperio nuclear and membrane IHC algorithms, for Ki-67 and CD3 staining, respectively. (Figure 2) These algorithms are based on the spectral differentiation between brown (positive) and blue (counter) staining. Total percentage positivity (1+ through 3+ added together) was recorded for each case.
Review of Pathology Report and Medical Chart
The percentage Ki-67 positivity was recorded from the original pathology report. If this number was given as a range (eg 70–80%), we used the mean (75% in the example given) for purposes of statistical analysis. Other clinical data collected included location of tumor, stage, international prognostic index (IPI) score, type of therapy received, date of diagnosis, outcome, date of death or last date of contact and overall survival time (defined as time from original diagnosis to death).
Statistical Analysis
Computer-generated quantitative image analysis (quantitative image analysis) percentages for Ki-67 were compared with the standardized value (mean percentage) extrapolated from the pathology report using Pearson correlations and paired t-tests. Pearson correlation with Ki-67 and CD3 was also calculated. Although quantitative image analysis CD3 values were also recorded, these were not typically mentioned in the pathology report, and thus no comparison could be made.
Patients were classified into low and high proliferation groups using cutoffs of 70 and 80%, and Kaplan–Meier survival curves were calculated for groups. Patients were censored at the end of the study. Log-rank tests were used to test whether survival curves were different between the two analysis modalities. Both the computer-generated and pathologist-estimated proliferation indices were compared with overall survival times, using both a 70 and 80% cutoff to stratify into a high and low proliferation group. Cox proportional hazards regression was used to further explore how proliferation index, age, gender and international prognostic index (IPI) score were associated with mortality. Cox models were used to assess whether CD3 was associated with mortality, and Ki-67 values were also compared with CD3 values to assess for any possible correlation.
Results
Patient characteristics are shown in Table 1. Of the 84 patients with valid follow-up times, 26 (31%) died of presumed lymphoma-associated complications. The median follow up time was 21 months (0.3–100 months).
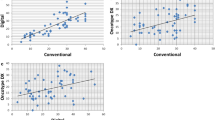
The pathologist estimate of Ki-67 percentages ranged 5 to 100%, with an average of 68.4% and a median of 77.5%. The computer estimate of Ki-67 percentage positivity ranged 7–96%, with an average of 62.0% and a median of 66.0%. Given that the pathologist-generated Ki-67 percentage is intended to be an estimate, assessment of the pathology reports reveals the expected finding that the pathologist is more likely to report the Ki-67 percentage in increments of 10 percentage points, while the computer-generated numbers are more continuous. Even with this difference, the pathologist estimate of proliferation index is still strongly correlated with the computer-generated value (P<0.001, r=0.67). Despite this strong correlation, the pathologist did tend to report a slightly higher value than did the computer (mean=6.0, 95% CI=2.0–10.1, P=0.004). We also noted poorer agreement at lower Ki-67 values. The majority of values were greater than 40% for both the pathologist and the computer, as would be expected in diffuse large B-cell lymphoma.
When Ki-67 values from both the pathologist and the computer were compared with survival outcomes, no statistically significant correlation was identified for either group. This included all diffuse large B-cell lymphoma cases taken together, as well as when subdivided by stage or international prognostic index (IPI). Although neither method was statistically significant, the pathologist estimate did show a trend of shorter survival with higher proliferation index (Figure 3).
In all, 65 cases had CD3 immunohistochemical stains of good quality available for quantitative image analysis. Of these, the percentage of CD3+ T-cells present in the specimen analyzed ranged 0–87%, with an average of 37% and a median of 40%.
We originally recorded CD3 percentages (computer generated) in each case to assess whether a higher number of background T-cells might in some way alter the computer's estimate of the Ki-67 proliferation index. The theory was that the computer would do a poorer job of separating true tumor cells from background non-neoplastic cells. Thus CD3 percentage was also compared with Ki-67 percentage and showed a negative correlation between the two variables (P<0.001), suggesting that CD3-positive cells did not stain positively for Ki-67. Interestingly, when isolated as an independent variable, CD3 percentage generated by the computer algorithm did show a statistically significant correlation with the overall survival, with higher percentages of CD3 associated with longer survival (P=0.014). (Figure 4) After adjusting for patient age and gender, the hazard of mortality decreased by 0.81 for each 10% increase in CD3 (P=0.037, 95% CI=0.67–0.99).
Discussion
Due to the tight correlation between quantitative image analysis and the pathologist assessment of Ki-67 percentage, at this point of time, there appears to be no particular benefit to using a computer-based algorithm, as the pathologist estimate is both accurate and adequate and has the benefit of not requiring additional equipment. As for the prognostic implications of the proliferation index in diffuse large B-cell lymphoma, our study was unable to demonstrate any significant correlation between proliferation rate and outcome. Although the pathologist estimate and survival outcome showed a trend of shorter overall survival with higher Ki-67 proliferation rate (using a cutoff of 80%), this finding was not statistically significant. The literature already shows conflicting data on this topic12, 13, 14, 15, 16, 17 and our findings do not lend any strong support to a correlation between the Ki-67 and outcomes.
Previous studies that showed strong association between Ki-67 proliferation rates and outcomes may have been affected by biases including deliberately selecting areas of highest proliferation and hand-counting tumor cells.19 We attempted to avoid these potential biases through the use of a computer-assisted algorithm, so as not to select only certain areas but to evaluate the tumor as a whole, encompassing intratumor variations in proliferation. Many previous studies were also performed on what are now outmoded chemotherapeutic regimens, namely CHOP without the addition of rituximab. In our study, the vast majority of patients completed R-CHOP chemotherapy. Our findings would suggest that either there is no correlation between proliferation rate and prognosis, or that possibly our sample size was too small to show a meaningful correlation. It would be worthwhile to have a large enough sample size to subdivide these outcomes into stage-dependent groupings. Unfortunately, although we attempted this type of analysis, our sample sizes were most likely too small to achieve a meaningful evaluation using these parameters.
Perhaps of greater interest was an unexpected finding in this study. Although we originally included the assessment of CD3 in this study, our intention was to use the percentage of T-cells to create a ‘correction factor’ so that the background T-cells would not falsely alter the proliferation rate. In the end, we decided against creating such a correction factor for two reasons: First, the pathologist estimate and computer estimate were already very closely aligned without the need for a correction factor. Second, comparison of the CD3 percentage and Ki-67 percentage from each case showed a negative correlation, suggesting that cells staining positively for CD3 did not, for the most part, stain positively for Ki-67 or contribute to the total Ki-67 percentage. This would suggest that background T-cells did not confound the Ki-67 percentage.
We then examined the CD3 percentage independently of the Ki-67 proliferation rate, with respect to its relationship with survival outcomes. CD3 is a pan T-cell antigen, and thus the CD3 percentage is an indicator of how many non-neoplastic T-cells are present in the background of the tumor. Diffuse large B-cell lymphomas typically do not express this marker and more specifically, none of our cases expressed CD3 in the tumor cells. It is also important to note that T-cell/histiocyte-rich large B-cell lymphomas were excluded from the study, as including this potentially aggressive lymphoma might confound our data. Our analysis showed a statistically significant correlation between increased numbers of T-cells and longer survival outcomes. We hypothesize that the recruitment of increased numbers of T-cells may indicate a more brisk or vigorous host inflammatory response, supported by the findings of Monti et al26 in their gene expression profiling study. If this is the case, this increased host response may prove beneficial in the survival outcomes of these patients.
Monti et al26 used molecular profiling to identify three distinct ‘signatures’ (gene expression profile patterns) in diffuse large B-cell lymphomas. One of these they coined the ‘Host Response’ signature, characterized by increased expression of T-cell receptor and inflammatory/immune components as well as a morphologically notable increase in T-cells, natural killer cells and macrophages. Although this group could be characterized as younger aged patients with frequent spleen and bone marrow involvement and fewer genetic abnormalities, there was no significant difference in survival between this and the other two groups.
Another study used flow cytometry to quantify the number of reactive T-cells in the tumor biopsy and found a correlation between CD4+ helper T-cells and overall survival.27 In contrast, Muris et al28 found that increased numbers of other groups of T-cells (activated cytotoxic T-lymphocytes expressing granzyme B) were associated with failure to reach complete remission and worse overall survival time. More work would still be needed in this area to further investigate, but from a practical standpoint, an assessment of background T-cells may be of use to the oncologist in prognostic assessment and biological behavior of the tumor.
For several years, the use of quantitative image analysis (quantitative image analysis) in diagnostic pathology has been a topic under considerable debate. In this study, we sought to investigate quantitative image analysis using Aperio ScanScope immunohistochemical algorithms for Ki-67 and CD3 stains versus original pathology reports, comparing Ki-67 percentages for the two methods with ultimate prognosis for patients diagnosed with diffuse large B-cell lymphoma.
Although there is significant correlation between pathologist estimates and quantitative image analysis of Ki-67 percentage, pathologists tend to report slightly larger values. Proliferation rates in diffuse large B-cell lymphoma have been associated with conflicting outcomes in the literature, and our data does not support that higher proliferation rates are association with worse clinical outcome. Interestingly, the Ki-67 percentage assessed by the pathologist may be more closely associated with survival outcome than that identified by quantitative image analysis. This may indicate that pathologists are better at selecting appropriate cells for analysis and avoiding the inclusion of background non-tumor cells in the estimate or that they are better able to evaluate the lymphoma sample within the context of the entire glass slide. Although a pathologist has the education and training to estimate the percentage positivity only from large tumor cells, effectively ignoring the smaller lymphocytes in the background, the degree of sophistication of the computer algorithm is likely still lacking and is less able to distinguish these subtle differences. Our study also suggests that quantitative image analysis may offer a way to quantify the presence of host inflammatory response, with results demonstrating that increased background (non-neoplastic) T cells (CD3 positive) are associated with improved survival.
Although more research is needed to evaluate potential clinical uses for quantitative image analysis in diagnostic pathology, our study shows that as there is substantial correlation between pathologist estimate and quantitative image analysis percentage for Ki-67-stained cells in diffuse large B-cell lymphoma, there is no benefit to using quantitative image analysis over pathologist estimate at this time. Further research must be done to investigate CD3 percentages as a possible prognostic variable in diffuse large B-cell lymphoma.
References
Stein H, Warnke RA, Chan WC, et al. In: Swerdlow SH, Campo E, Harris NL, et al. (eds). Diffuse large B-cell lymphoma, not otherwise specified. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC: Lyon, 2008, pp 233–237.
Saez AI, Saez AJ, Artiga MJ, et al. Building an outcome predictor model for diffuse large B-cell lymphoma. Am J Pathol 2004;164:613–622.
Shipp MA, Ross KN, Tamayo P, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med 2002;8:68–74.
Tagawa H, Suguro M, Tsuzuki S, et al. Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood 2005;106:1770–1777.
Lossos IS, Czerwinski DK, Alizadeh AA, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med 2004;350:1828–1837.
Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–511.
Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004;103:275–282.
Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large B-cell lymphoma. N Engl J Med 2002;346:1937–1947.
Silvestrini R, Costa A, Boracchi P, et al. Cell proliferation as a long-term prognostic factor in diffuse large-cell lymphomas. Int J Cancer 1993;54:231–236.
Mounier N, Brier J, Gisselbrecht C, et al. Rituximab plus CHOP (R-CHOP) overcomes bcl-2-associated resistance to chemotherapy in elderly patients with diffuse large B-cell lymphoma (diffuse large B-cell lymphoma). Blood 2003;101:4279–4284.
Rimsza LM, Leblanc ML, Unger JM, et al. Gene expression predicts overall survival in paraffin-embedded tissues of diffuse large B-cell lymphoma treated with R-CHOP. Blood 2008;112:3425–3433.
Broyde A, Boycov O, Strenov Y, et al. Role and prognostic significance of the Ki-67 index in non-Hodgkin's lymphoma. Am J Hematol 2009;84:338–343.
Sanchez E, Chacon I, Plaza MM, et al. Clinical outcome in diffuse large B-cell lymphoma is dependent on the relationship between different cell-cycle regulator proteins. J Clin Oncol 1998;16:1931–1939.
Miller TP, Grogan TM, Dahlberg S, et al. Prognostic significance of the Ki-67-associated proliferative antigen in aggressive non-Hodgkin's lymphomas: a prospective Southwest Oncology Group trial. Blood 1994;83:1460–1466.
Colomo L, Lopez-Guillermo A, Ojanguren J, et al. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood 2003;101:78–84.
Zhang A, Ohshima K, Sato K, et al. Prognostic clinicopathologic factors, including immunologic expression in diffuse large B-cell lymphomas. Pathol Int 1999;49:1043–1052.
Hasselblom S, Ridell B, Sigurdardottir M, et al. Low rather than high Ki-67 protein expression is an adverse prognostic factor in diffuse large B-cell lymphoma. Leuk Lymph 2008;49:1501–1509.
Ross W, Hall PA . Ki-67: from antibody to molecule to understanding? J Clin Pathol Mol Pathol 1995;48:M113–M117.
Grogan TM, Lippman SM, Spier C, et al. Independent prognostic significance of a nuclear proliferation antigen in diffuse large cell lymphomas as determined by the monoclonal antibody Ki-67. Blood 1988;71:1157–1160.
Gilbertson JR, Ho J, Anthony L, et al. Primary histologic diagnosis using automated whole slide imaging: a validation study. BMC Clin Pathol 2006;6:4.
Fine JL, Grzybicki DM, Silowash R, et al. Evaluation of whole slide image immunohistochemistry interpretation in challenging prostate needle biopsies. Hum Pathol 2008;39:564–572.
Furness P . A randomized controlled trial of the diagnostic accuracy of internet-based telepathology compared with conventional microscopy. Histopathology 2007;50:266–273.
Lopez C, Lejeune M, Salvado MT, et al. Automated quantification of nuclear immunohistochemical markers with different complexity. Histochem Cell Biol 2008;129:379–387.
Rexhepaj E, Brennan DJ, Holloway P, et al. Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: application to measurement of oestrogen and progesterone receptor levels in breast cancer. Breast Cancer Res 2008;10:R89.
Kanungo A, Medeiros LJ, Abruzzo LV, et al. Lymphoid neoplasms associated with concurrent t(14;18) and 8q24/c-MYC translocation generally have a poor prognosis. Mod Pathol 2006;19:25–33.
Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005;105:1851–1861.
Ansell SM, Stenson M, Habermann TM, et al. CD4+ T-cell immune response to large B-cell non-Hodgkin's lymphoma predicts patient outcome. J Clin Oncol 2001;19:720–726.
Muris JJF, Meijer CJLM, Cillessen SAGM, et al. Prognostic significance of activated cytotoxic T-lymphocytes in primary nodal diffuse large B-cell lymphomas. Leuk 2004;18:589–596.
Acknowledgements
We would like to thank the Foucar Endowment Fund for generously providing a grant to complete this research project. This study was also partially supported by DHHS/NIH/NCRR Grant #1UL1RR031977-01, The University of New Mexico Clinical and Translational Science Center.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Chabot-Richards, D., Martin, D., Myers, O. et al. Quantitative image analysis in the assessment of diffuse large B-cell lymphoma. Mod Pathol 24, 1598–1605 (2011). https://doi.org/10.1038/modpathol.2011.123
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2011.123
Keywords
This article is cited by
-
Computer-driven quantitative image analysis in the assessment of tumor cell and T cell features in diffuse large B cell lymphomas
Annals of Hematology (2018)
-
Persistent homology index as a robust quantitative measure of immunohistochemical scoring
Scientific Reports (2017)
-
From drug discovery to biomarker-driven clinical trials in lymphoma
Nature Reviews Clinical Oncology (2012)