Abstract
The local immune response occurring during Staphylococcus aureus nasal colonization remains ill-defined. Studies have highlighted the importance of T-cell immunity in controlling S. aureus colonization of the nasal mucosa. We extend these observations, identifying a critical role for interleukin (IL)-22 in this process. IL-22 is basally expressed within the nasal mucosa and is induced upon S. aureus colonization. IL-22 is produced by CD4+ and CD8+ T lymphocytes at this site, with innate-like lymphocytes also contributing. IL-22−/− mice demonstrate significantly elevated levels of S. aureus nasal colonization as compared with wild-type (WT) mice. This was associated with reduced expression of antimicrobial peptides (AMPs) in the nose. Furthermore, expression of staphylococcal ligands loricrin and cytokeratin 10 was higher in the noses of IL-22−/− as compared with WT mice. IL-17 has been shown to regulate S. aureus nasal colonization by controlling local neutrophil responses; however, IL-17 expression and neutrophil responses were comparable in the noses of IL-22−/− and WT mice during S. aureus colonization. We conclude that IL-22 has an important role in controlling S. aureus nasal colonization through distinct mechanisms, with IL-22 mediating its effect exclusively by inducing AMP expression and controlling availability of staphylococcal ligands.
Similar content being viewed by others
Introduction
Staphylococcus aureus is an opportunistic pathogen, responsible for several life-threatening invasive diseases, as well as less severe skin and soft tissue infections. In contrast to its invasive ability, S. aureus is part of the normal human microbiome and persistently colonizes the anterior nares of 20% of the population, whereas the remainder are colonized intermittently.1 The anterior nares are the principal niche for S. aureus and often act as a reservoir for the inoculation of other body sites.2 Persistent carriage is a risk factor for invasive infection particularly in a nosocomial setting and patients are frequently infected by their endogenous strain.3 Temporary decolonization of the nose is achieved in at-risk patients using topical antibiotics such as mupirocin; however, effectiveness of this strategy is hampered by escalating mupirocin resistance.4 A number of studies have begun to elucidate the molecular interactions that occur between the organism and the host at the surface of the nasal epithelium;5, 6 however, the immune response at this site during S. aureus colonization remains largely undefined. A detailed understanding of local immunity at the primary carriage site of S. aureus may provide the framework for the development of therapeutic strategies to enhance the host immune response in order to effectively eradicate bacterial nasal carriage in target patient groups.
Although S. aureus is primarily found in the anterior nares, clinical studies support the presence of several sites of colonization within the deeper areas of the nasal cavity, as well as the throat.7, 8 The mechanisms underlying the interaction between S. aureus and the nasal epithelium have been defined for two sites within the nasal cavity: the anterior nares and epithelial cells lining the inner nasal cavity.5, 6 Bacterial attachment to the anterior nares is facilitated by the staphylococcal surface adhesin clumping factor B, through high-affinity interaction with the cornified envelope (CE) protein loricrin.6 Colonization of the inner nasal cavity is mediated by the cell wall glycopolymer wall teichoic acid. Wall teichoic acid binds to an F-type scavenger receptor, known as scavenger receptor expressed by endothelial cell-1 (SREC-1), which is expressed on the surface of epithelial cells in the inner nasal cavity.5 Given that the desquamated epithelial cells to which S. aureus binds at the anterior nares are constantly shed from the nose, it has been proposed that the colonization site at the inner nasal epithelium acts as a reservoir of bacteria for the anterior nares.5
A number of genetic determinants of S. aureus nasal carriage have been reported. Polymorphisms in genes expressing human β-defensin 1, mannose-binding lectin, C-reactive protein, and IL-4 are associated with an increased risk for S. aureus nasal carriage,9, 10, 11 highlighting that S. aureus cultivates a unique relationship with both the innate and the adaptive immune system during nasal colonization. Nasal fluid from non-carriers displays stronger antimicrobial activity against S. aureus when compared with nasal secretions from carriers7 and impaired induction of the human antimicrobial peptide (AMP) β defensin 3 in the skin correlates with persistent carriage status.12 Clinical observations have revealed the importance of adaptive immunity. Defined non-carriers and intermittent carriers have similar antistaphylococcal antibody profiles against S. aureus antigens that are distinct from persistent carriers,1 and persistent carriage is linked with higher levels of antistaphylococcal IgA and IgG in children.13 Furthermore, an increased prevalence of S. aureus colonization is observed in HIV patients with lower CD4+ T-cell counts, suggesting that T cells have an important role in controlling S. aureus colonization.14 To date, only a single murine study has been undertaken to dissect this response at the cellular level and has identified that T-helper type 17 (Th17) cells are important in controlling S. aureus nasal colonization through their ability to promote neutrophil recruitment to the anterior nares.15
Like the intestinal mucosa, the nasal mucosa is continually exposed to a diverse ecosystem of microorganisms, necessitating a multi-factorial immune response to defend against inhaled pathogens and control the commensal microbiota. The cytokine IL-22 has emerged as a key player in intestinal host defense through its ability to promote production of AMPs16 and to promote tissue healing.17 IL-22 mediates the expression of many AMPs in the gut, skin, and lungs, including β-defensins 2 and 3, S100A7-9, and Reg3γ.16, 18, 19, 20, 21 IL-22 is required to prevent severe intestinal pathology and mortality during Clostridium rodentium colitis and is critical for host protective immunity against Klebsiella pneumoniae in the lung.16, 19 In addition to its role in regulating AMP production, IL-22 inhibits the terminal differentiation of keratinocytes by downregulating expression of CE proteins, such as loricrin and cytokeratin 10 (K10), and induces keratinocyte migration to promote wound healing in the epithelium.21, 22
IL-22 is primarily produced by conventional T cells, γδT cells, and innate lymphoid cells (ILCs). Studies using an IL-22 fate reporter mouse have indicated that γδT cells are the predominant source of IL-22 in the mouse skin and lungs, whereas type-3 ILCs are the primary source of IL-22 in the intestine.23 In humans, a distinct subset of skin-homing memory CD4+ T cells, termed Th22 cells, are a major source of IL-22 (ref. 24) and IL-22 can also be produced by the CD8+ counterpart Tc22 at this site.25 Furthermore, a subset of mucosal-associated natural killer cells, termed NK22 cells, in the tonsils and payers patches exclusively produce IL-22 and may have a role in innate immunity at mucosal sites.26 Type-3 ILCs have emerged as key orchestrators of immune defense and homeostasis in the gut27 but have also been shown to be responsible for IL-22 production in a murine model of Streptococcus pneumoniae lung infection.28 γδT cells have been identified as an important source of IL-22 during Bacillus subtilis lung infection29 and C. rodentium intestinal infection,30 and there is emerging evidence that IL-22 derived from CD8 cells is important in protection against oropharyngeal candidiasis.31 Recently, IL-22 produced by both conventional TCRβ+ cells and also a population of CD3− cells, potentially ILCs, was shown to be protective in S. aureus pneumonia infection.32
In this study, we define a role for IL-22 in the host response to S. aureus nasal colonization. We demonstrate that IL-22 expression is induced in a number of lymphocyte populations within the nasal mucosa following colonization by S. aureus. Mice deficient in IL-22 were more readily colonized with S. aureus and this was associated with impaired AMP production locally within the nasal cavity. Furthermore, we demonstrate for the first time that expression of known ligands for S. aureus attachment in the nose is controlled in vivo by IL-22. Our findings identify IL-22 as one of the critical components of the host immune response that controls nasal colonization by S. aureus, mediated through its ability to induce killing of the organism by triggering AMP expression, and importantly by inhibiting attachment to the nares by controlling availability of staphylococcal ligands.
Results
IL-22 is constitutively expressed in nasal tissue and is induced upon S. aureus colonization
To establish whether IL-22 is basally expressed in the nasal tissue, RNA was isolated from the skin, spleen, kidney, lungs, nose, and nasopharyngeal tissue (NT) of naive WT mice and IL-22 messenger RNA (mRNA) expression was determined using quantitative real-time PCR (qPCR). Basal expression of IL-22 was significantly elevated in the nose when compared with the spleen and kidney (Figure 1a). High IL-22 expression was also observed in the NT with some basal expression detected in the skin. IL-17 expression followed a similar pattern with basal expression of IL-17 detectable in the skin, nose, and NT (Figure 1b). Constitutive expression of IL-22 and IL-17 in response to the commensal microbiome in the gut has been reported;27, 33, 34 our findings indicate that IL-22 and IL-17 may also be expressed locally at nasal mucosa.
Basal expression of interleukin (IL)-22 and IL-17 in murine nasal tissue. Naive WT mice were killed and RNA was extracted from the spleen, kidney, lungs, nose, and nasopharynx-associated lymphoid tissue (NALT). IL-22 (a) and IL-17 (b) gene expression was assessed using quantitative reverse transcription PCR. Messenger RNA (mRNA) values were expressed as mean relative expression ±s.e.m. compared with 18S RNA expression (n=7–9, per group). Statistical analysis was performed using a Kruskal–Wallis test. *P<0.05; **P<0.005.
A previous study has shown that IL-17 is induced in the nasal cavity during S. aureus colonization.15 In order to establish whether IL-22 is also induced in response to S. aureus colonization of the nasal cavity, S. aureus nasal colonization was established in the groups of WT mice using a spontaneous streptomycin-resistant mutant of S. aureus strain Newman (Newman SmR, 2 × 108 colony-forming unit (CFU) per nose). Over the course of 14 days, IL-22 mRNA and protein levels were determined in the nose and NT of colonized WT mice and compared with the baseline IL-22 expression levels in non-colonized phosphate-buffered saline (PBS)-treated controls. IL-22 mRNA expression was significantly upregulated at both sites by day 7 post colonization (Figure 2a,b). Significant levels of IL-22 protein (above PBS control levels) were detectable in the nose on days 3 and 10 post colonization (Figure 2c).
Expression of interleukin (IL)-22 in the nasal cavity following S. aureus colonization. Wild-type mice were colonized with S. aureus Newman SmR (2 × 108 colony-forming units per nose) or were administered with phosphate-buffered saline (PBS). On days 1, 3, 7, 10, and 14 mice were killed, and RNA was extracted from the nares and nasopharyngeal tissue (NT). Analysis of IL-22 gene expression in the nares (a) and NT (b) was established using quantitative reverse transcription PCR. Messenger RNA values were expressed as mean fold increase in expression ±s.e.m. and was compared with baseline IL-22 expression from PBS-treated controls after normalizing to 18S RNA expression (n=5–6, per group). On days 1, 3, and 10, noses were homogenized in PBS and protein levels of IL-22 (c) were determined by enzyme-linked immunosorbent assay. Values are expressed as mean protein concentration ±s.e.m. (n=5, per group). Statistical analysis was performed using two-way analysis of variance or a Kruskal–Wallis test with Dunns Multiple Comparisons. *P≤0.05; **P<0.005; ***P≤0.001.
IL-22 is produced by CD4+ and CD8+ T cells as well as a population of ILCs in the nasal tissue during S. aureus nasal colonization
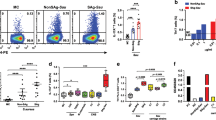
To establish the cellular source of IL-22 in the nasal cavity during S. aureus nasal colonization, mice were colonized with Newman SmR, and 7 days post colonization, the NT (comprising the nasopharynx-associated lymphoid tissue) was excised for the analysis of leukocyte populations by flow cytometry combined with intracellular cytokine staining for IL-22. Following S. aureus nasal colonization, total numbers of CD45+ leukocytes were increased twofold in the NT, confirming that an immune response is activated (Figure 3a). Of these CD45+ leukocytes, ∼3% was IL-22 producing, whereas IL-22 expression was undetectable (<0.5%) in non-colonized control mice (Figure 3b). To establish the cellular source of IL-22, we used a panel of well-defined surface markers to identify individual T-cell subsets. On day 7 post colonization, IL-22 was predominantly produced by CD4+ T cells (CD3+CD4+) and CD8+ T cells (CD3+CD8+). IL-22 was also produced by a population of CD3−CD4+ cells, which potentially represent type-3 ILCs (Figure 3c). However, the low numbers of these cells recoverable from the NT (∼200 cells) precluded further phenotypic analysis. We did not detect IL-22-producing γδ T cells (CD3+TCRδ+) at this time point.
Source of IL-22 in the nasopharyngeal tissue (NT) during S. aureus nasal colonization. Wild-type mice were colonized with S. aureus Newman SmR (2 × 108 colony-forming units per nose) or were administered with phosphate-buffered saline (PBS). At day 7 post colonization, mice were killed and cells were isolated from the NT. Cells were cultured with brefeldin A and were surface stained with CD45, CD3, CD4, CD8, and TCRδ before intracellular staining for interleukin (IL)-22 and analysis by flow cytometry. Total infiltrating of CD45+ leukocytes (a) was assessed and a significant proportion was shown to produce IL-22 (b). Analysis of IL-22 production by individual T-cell subsets was then assessed (c). Results are expressed as mean±s.e.m. with representative fluorescence-activated cell sorting plots shown (n=4–5, per group). Data are representative of two individual experiments. SSC, side scatter.
S. aureus nasal colonization is elevated in IL-22-deficient mice
Having demonstrated that IL-17 and IL-22 are constitutively expressed in the nasal cavity, we used IL-17- and IL-22-deficient mice to examine the role of these cytokines in controlling S. aureus nasal colonization. WT, IL-17A−/−, and IL-22−/− mice were inoculated intra-nasally with Newman SmR (2 × 108 CFU per nose) and bacterial burdens in the noses enumerated on days 10 and 14 post colonization. S. aureus nasal colonization levels were significantly elevated in IL-22−/− and IL-17A−/− mice on day 14 as compared with WT mice (Figure 4a). Similarly, although not statistically significant, IL-22−/− and IL-17−/− mice retained higher levels of bacteria in their noses compared with WT mice on day 10 post colonization. These results confirm previous studies that suggest that IL-17 is important for clearance of S. aureus from the nasal cavity, and indicate that IL-22 may also have an important role in controlling S. aureus persistence at this site.
Bacterial burden is significantly elevated in the nasal cavity interleukin (IL)-22-deficient mice during S. aureus colonization. Groups of wild-type (WT), IL-17A−/−, and IL-22−/− mice were colonized using S. aureus Newman SmR (2 × 108 colony-forming units (CFU) per nose). At days 3, 10, 14, and 21, mice were killed, and bacterial burden in the noses (a, b) and nasopharyngeal tissue (d) was established. Results expressed as mean CFU per nose (n=10–15, per group). Colonization rate (c) as determined by the number of mice colonized by S. aureus/total number of mice was determined in WT and IL-22−/− mice (n=10–15, per group). Statistical analysis was performed using a Kruskal–Wallis test. *P<0.05; **P<0.005; ***P<0.0001.
We then examined S. aureus nasal colonization in IL-22−/− mice over the course of 21 days. On day 3 post colonization, both WT and IL-22−/− mice retained similar numbers of S. aureus in the nose (Figure 4b). However, by day 10, IL-22−/− mice had a higher bacterial burden providing evidence of S. aureus proliferation in the nose when compared with WT mice. By day 14, bacterial burden in IL-22−/− mice was significantly elevated when compared with WT mice (P=<0.05) and after 21 days, low levels of S. aureus remained in the noses of IL-22−/− mice, whereas S. aureus was completely cleared from WT mice. The carriage rate in the nose (determined by the number of mice colonized by S. aureus/total number of mice) in WT mice decreased to 50% at day 14 post colonization, whereas IL-22−/− mice maintained 100% carriage rate at this time point, and by day 21, WT mice displayed 100% clearance compared with only 60% clearance rate seen in IL-22−/− mice (Figure 4c).
To assess the ability of S. aureus to survive in the NT, nasopharyngeal lavages were performed (following removal of the nose) on WT and IL-22−/− mice post colonization and the bacterial burden was assessed. Overall, IL-22−/− mice retained more bacteria in the nasopharyngeal lumen than WT mice (Figure 4d). On day 3 post inoculation, there was a significantly higher bacterial burden obtained from IL-22−/− mice compared with WT mice.
To assess the therapeutic effect of IL-22 in the nose, recombinant murine IL-22 (200 ng per nose) was administered to WT and IL-22−/− mice in conjunction with S. aureus intranasal inoculation. On day 3 post colonization, WT mice that received rIL-22 exhibited a significant reduction in colonization compared with WT mice treated with PBS (Figure 5a). Similarly, administration of rIL-22 to IL-22−/− mice significantly reduced S. aureus colonization (Figure 5b). Taken together, these data identify IL-22 as one of the major factors controlling S. aureus colonization of the nasal cavity.
Treatment with recombinant IL-22 reduces bacterial burden during S. aureus nasal colonization. Groups of wild-type (a) and IL-22−/− (b) mice were colonized using S. aureus Newman SmR (2 × 108 colony-forming units (CFU) per nose) with or without recombinant murine IL-22 added (200 ng per nose). At day 3 post colonization, mice were killed and bacterial burden in the noses was established. Results expressed as mean CFU per nose (n=10, per group). Statistical analysis was performed using a Mann–Whitney test. *P<0.05.
IL-22 does not regulate the IL-17 axis or neutrophil recruitment during nasal colonization
IL-17-dependent recruitment of neutrophils to the nasal cavity has proven important for controlling S. aureus nasal colonization.15 Therefore, it was important to establish whether IL-22 may in-part mediate its effect through IL-17 induction and associated neutrophil recruitment. A 14-day time course of S. aureus nasal colonization was established in the groups of WT and IL-22−/− mice, and at specific time points post colonization expression of inflammatory cytokines and chemokines in the nose was assessed. Consistent with previous studies, an increase in the mRNA expression of IL-17 and CXCL1 was observed as early as day 1 post colonization in WT mice. IL-17 and CXCL1 mRNA, and protein levels were elevated in the first 7 days post colonization (Figure 6a–d). Although CXCL1 mRNA levels appeared to be higher in WT mice, no significant difference in expression of either IL-17A or CXCL1 mRNA or protein was observed between WT and IL-22−/− mice during colonization. IL-1β is known to be critically important in driving IL-17 and IL-22 responses.30, 35, 36 To establish whether IL-22 had any effects on IL-1β activity during S. aureus nasal colonization, IL-1β protein levels were assessed in the nose. IL-1β was elevated at day 1, but by day 7, a decline in expression was observed in both WT and IL-22−/− animals (Figure 6e).
Cytokine expression in the nares of wild-type (WT) and interleukin (IL)-22−/− mice during S. aureus nasal colonization. Groups of WT and IL-22−/− mice were colonized using S. aureus Newman SmR (2 × 108 colony-forming units per nose) or were administered with phosphate-buffered saline (PBS). At days 1, 3, 5, 7, 10, and 14 mice, were killed, RNA was extracted from noses, and gene expression of IL-17A (a) and CXCL1 (c) assessed using quantitative reverse transcription PCR. Expression was compared with PBS controls after normalizing to 18S RNA expression. Values are expressed as mean fold increase compared with PBS controls ±s.e.m. (n=7–9, per group). For analysis of protein, noses were homogenized in PBS and concentration of IL-17 (b), CXCL1 (d), and IL-1β (e) determined by enzyme-linked immunosorbent assay. Values are expressed as mean protein concentration ±s.e.m. (n=7–10, per group).
Neutrophil infiltration to the nose and the NT was also assessed in WT and IL-22−/− mice on day 7 post colonization. Although neutrophil recruitment to both the nose and NT was increased following S. aureus colonization compared with PBS controls, neutrophil recruitment to either site was comparable in both WT and IL-22−/− mice (Figure 7). Similarly, neutrophil numbers did not differ between WT and IL-22−/− mice on day 3 post colonization (Supplementary Figure S1 online). These results therefore suggest that in contrast to IL-17, IL-22 does not control S. aureus nasal colonization by regulating the local neutrophil response.
Neutrophil influx to the nasal cavity of wild-type (WT) and interleukin (IL)-22−/− mice during S. aureus nasal colonization. Groups of WT and IL-22−/− mice were colonized using S. aureus Newman SmR (2 × 108 colony-forming units per nose) or were administered with phosphate-buffered saline (PBS). Cells isolated from the nose and nasopharyngeal tissue (NT) on day 7 post colonization were stained with anti-CD11b, anti-Ly6G, and anti-F4/80 and analyzed by flow cytometry. Neutrophils were identified as CD11b+ Ly6G+ F4/80− cells. Values are expressed as mean absolute numbers of neutrophils isolated from nasal tissue ±s.e.m. (n=4–5, per group). Data are representative of two different experiments.
The expression of AMP is reduced in IL-22−/− mice
IL-22 is a potent inducer of AMP expression during bacterial infection at the lung and gut epithelium.16, 19 To determine the role of AMPs in IL-22-mediated protection against S. aureus nasal colonization, noses from WT and IL-22−/− mice were analyzed for AMP expression using qPCR. The expression of murine β-defensins 3, 4, and 14, (Figure 8a–c), Reg3γ (Figure 8d), S100A8, and S100A9 (Figure 8e,f) was determined in the nose of WT and IL-22−/− mice compared with mice inoculated with PBS.
Differential expression of AMPs in wild-type (WT) and interleukin (IL)-22−/− mice during S. aureus nasal colonization. Groups of WT and IL-22−/− mice were colonized using S. aureus Newman SmR (2 × 108 colony-forming units per nose) or were administered with phosphate-buffered saline (PBS). At days 1, 3, 7, 10, and 14, mice were killed, RNA was extracted from noses, and gene expression of defβ3 (a), defβ4 (b), defβ14 (c), regIIIγ (d), S100A8 (e), and S100A9 (f) in the noses assessed using quantitative reverse transcription PCR. Expression was compared with PBS controls after normalizing to 18s RNA expression and expressed as mean fold increase in expression ±s.e.m. (n=3, per group). Data are representative of at least two individual experiments. Statistical analysis was performed using two-way analysis of variance. *P<0.05; **P<0.005; ***P<0.001.
In WT mice, expression of β-defensins 3, 4, Reg3γ, and S100A9 was upregulated at the early stages of colonization, whereas expression of β-defensin 14 and S100A8 peaked later. The expression of all AMPS was almost completely abolished in IL-22−/− mice. These results indicate that a spectrum of AMPs is produced in the nose at different stages in response to S. aureus nasal colonization, and that their production is dependent upon IL-22. Interestingly, there was no significant change in AMP expression in the noses of IL-17A−/− mice compared with WT mice at days 1, 3 and 7 post colonization (Supplementary Figure S2).
Expression of squamous cell epithelium staphylococcal ligands is more abundant in the nares of IL-22−/− mice
Previous studies have reported that IL-22 downregulates the expression of many structural proteins in the CE of epithelial cells.21, 22 Two of these proteins, loricrin and K10, have been implicated as ligands for staphylococcal adhesion to desquamated epithelial cells.6, 37 To determine whether a deficiency of IL-22 affects expression of loricrin and K10 in the nares, RNA extracted from nasal homogenates was probed for the expression of loricrin and K10 using qPCR. IL-22−/− control mice displayed higher basal expression of loricrin and K10 (Figure 9a,b) compared with WT control mice. In WT mice, both loricrin and K10 mRNA expression levels remained similar to PBS controls 1 and 3 days post colonization. However, loricrin and K10 levels were increased on days 1 and 3 post colonization in IL-22−/− mice as compared with the PBS controls and were significantly elevated as compared with WT colonized mice.
Expression of loricrin and cytokeratin 10 in the nares of wild-type (WT) and interleukin (IL)-22−/− mice. Groups of WT and IL-22−/− mice were colonized using S. aureus Newman SmR (2 × 108 colony-forming units per nose) or phosphate-buffered saline (PBS). At days 1 and 3, mice were killed, RNA was extracted from noses, and gene expression of loricrin (a) and K10 (b) assessed using quantitative reverse transcription PCR. Messenger RNA values were expressed as mean fold increase compared to PBS-treated controls after normalizing to 18S RNA expression ±s.e.m. (n=7–8, per group). Statistical analysis was performed using Mann–Whitney test. Nasal tissue was excised from euthanized mice at days 1 and 3 post colonization. Soluble proteins were extracted and analyzed by western immunoblotting using rabbit anti-murine loricrin IgG (c, e) or rabbit anti-murine K10 IgG (d, e). The band intensity of samples was measured using ImageQuant software. β-Actin expression was used as a loading control. Values are expressed as ratio of mean expression compared with β-actin ±s.e.m. (n=10–12, per group). Statistical analysis was performed using two-way analysis of variance. *P<0.05; **P<0.005.
To confirm this at the protein level, protein extracts from the noses of WT and IL-22−/− mice were probed for loricrin and K10 on days 1 and 3 post colonization compared with PBS controls using western immunoblotting. Nasal homogenates were normalized by protein concentration, and loricrin and K10 were detected as bands of ∼56 and 60 kDa, respectively. On day 1 post colonization, loricrin (Figure 9c,e) and K10 (Figure 9d,e) protein expression following S. aureus nasal colonization in WT mice was not significantly altered from PBS controls. Consistent with this, we previously observed no significant difference in loricrin expression after 10 days of S. aureus nasal colonization in WT mice.6 In contrast, on day 1 post colonization in IL-22−/− mice, loricrin and K10 protein expression was higher compared with control mice and was significantly higher than loricrin and K10 expression in WT mice at this time point. This trend was still evident on day 3 post colonization. Taken together, these results indicate that during S. aureus nasal colonization induction of IL-22 locally in the nasal cavity controls expression of squamous epithelium ligands. This change in ligand-availability may affect S. aureus persistence.
Discussion
There is currently a lack of understanding of how the host immune response controls nasal colonization with S. aureus. There is evidence to suggest that Th17-mediated responses are important for clearance of S. aureus by regulating neutrophil recruitment to the nose.15, 38 Our study now demonstrates a critical role for IL-22 in controlling S. aureus nasal colonization. We demonstrate enhanced expression of IL-22 in the nose during nasal colonization with S. aureus, and that this cytokine is produced by innate and adaptive lymphocytes. IL-22 regulates the production of AMPs locally within the nose and importantly controls expression of staphylococcal-binding ligands on the CE of cells in the nose. Through these effects, IL-22 contributes to the elimination of S. aureus from the nasal cavity. Furthermore, administration of rIL-22 to the nose may have therapeutic benefit for nasal decolonization of S. aureus.
IL-22 is expressed basally within the nasal cavity of naive mice and is induced following colonization with S. aureus. Analogous to IL-22 expression observed in the gut mucosa,27, 34 IL-22 is likely constitutively expressed at the nasal mucosa in response to commensal nasal flora. Consistent with this, we observed that treatment with streptomycin, which reduced the total nasal flora (Supplementary Figure S3), was associated with reduced IL-22 expression in the nose. Moreover, IL-22-producing cells were undetectable in the NT of non-colonized (streptomycin treated) mice. Germ-free mice display lower levels of IL-22-producing innate immune cells and IL-22 mRNA in the gut.27, 34 Our findings are consistent with this and indicate that IL-22 production is induced by commensal nasal flora. Basal IL-17 expression is also high in the nose compared with other tissues, suggesting that IL-17 is also expressed in the nares in response to commensal flora, a mechanism also seen previously in germ-free mice.33 These data indicate that IL-22 is a key player in modulating microbial homeostasis at the nasal mucosal surface.
Previous studies of S. aureus and S. pneumoniae colonization have utilized mice deficient in leukocyte subsets to identify that a T-cell-mediated response is required for bacterial clearance from the nose and nasopharynx.15, 38 Until now, no study has attempted to phenotype T-cell subsets expanded locally within the nasal cavity in response to bacterial colonization. We observed that CD4+ and CD8+ T cells, and to a lesser extent a population of cells that are likely ILCs, are expanded in the nasal cavity during S. aureus nasal colonization (Supplementary Figure S4A). IL-22 was predominantly produced by CD4+ and CD8+ cells, whereas ILCs also potentially contribute to IL-22 production. It was recently shown that TCRβ+ cells were the predominant source of IL-22 in a S. aureus pneumonia model, whereas IL-22 production was localized to CD4+ and CD8+ T cells in the lesional skin in a murine model of atopic dermatitis. These cells exclusively produce IL-22 in the absence of IL-17, alluding to the presence of specialized “T22” T-cell subsets in the skin.39 In this study, we did not specifically determine whether IL-22-producing T cells co-synthesized IL-17; however, we were unable to detect IL-17 by fluorescence-activated cell sorting on day 7 post colonization (Supplementary Figure S4B). Archer et al. demonstrated that severe combined immunodeficiency mice retained elevated CFU in the nares compared with IL-17A−/− mice at 28 days post colonization, suggesting that other Th17 cytokines are involved in S. aureus nasal clearance.15 Both IL-17A and IL-17F have been shown to confer protection against S. aureus murine mucocutaneous infection;40 however, our data implicate that in the nasal cavity, IL-22 is a critical factor in response to S. aureus colonization. A role for IL-17F in controlling S. aureus nasal colonization remains to be established.
The antimicrobial properties of nasal fluid have been implicated as a determining factor of S. aureus carriage status. Studies using human keratinocytes show that expression of β-defensins 2 and 3 is induced in response to S. aureus, and β-defensin 3 displays potent antistaphylococcal activity.41 Reg3γ is both bacteriostatic and bactericidal against S. aureus due to its ability to bind peptidoglycan.42 Using qPCR, we observed a significant increase in AMP expression in the nares following S. aureus nasal colonization that was absent in IL-22−/− mice. Our results demonstrate that AMP expression correlates with S. aureus nasal colonization, and that IL-22 is a critical factor regulating their expression. IL-22 has been shown to synergistically enhance IL-17-mediated production of β-defensin 2, S100A7, S100A8, and S100A9 in human keratinocytes.18 However, we observed no significant change in AMP expression in the noses of IL-17A−/− compared with WT mice following S. aureus colonization, indicating that IL-22 has a dominant role in controlling AMP expression at this site.
IL-22 can inhibit terminal differentiation of keratinocytes by downregulating expression of several proteins in the CE of keratinocytes, including K10 and loricrin.21, 22 Loricrin, which comprises ∼80% of the CE of terminally differentiated keratinocytes, is a determinant of S. aureus nasal colonization. Loricrin interacts specifically with the S. aureus surface protein clumping factor B mediating attachment to murine nares and to human terminally differentiated keratinocytes.6 K10 also mediates S. aureus attachment to terminally differentiated keratinocytes in a clumping factor B-dependent mechanism.37 Here we show, for the first time, a host defense mechanism that controls availability of staphylococcal ligands in vivo. Our data support a mechanism whereby upon S. aureus colonization, IL-22 is induced and manipulates a keratinocyte response pathway, resulting in inhibition of terminal keratinocyte differentiation. In IL-22−/− mice, the absence of this pathway may result in more terminally differentiated, loricrin- and K10-rich cells, and could facilitate greater attachment of S. aureus to the nares. IL-22-signaling in keratinocytes is primarily mediated by the transcription factor STAT3.22 Activation of STAT3 by IL-22 and subsequent repression of keratinocyte differentiation has been demonstrated in vitro.43 Furthermore, STAT3 activation and STAT3-mediated Reg3γ expression are crucial for clearance of meticillin-resistant S. aureus (MRSA) pneumonia in vivo,44 but whether this mechanism is involved during S. aureus nasal colonization remains to be elucidated. In an in vitro keratinocyte culture system, induction of IL-6 by S. aureus triggers the downregulation of loricrin and K10.45 However, similar levels of IL-6 were induced in the noses of WT and IL-22−/− mice after 1 day of S. aureus colonization (Supplementary Figure S5), suggesting that IL-22 is regulating expression of loricrin and K10 in the nose directly, and that these effects are not mediated by IL-6 in vivo.
This study outlines a mechanism by which IL-22 regulates S. aureus nasal colonization that is distinct from IL-17. We hypothesize that IL-17 and IL-22 work in concert to control S. aureus nasal colonization, but operate through distinct mechanisms; IL-22 regulates bacterial burden through the induction of AMPs and by controlling keratinocyte differentiation and thus availability of S. aureus ligands within the nose. In contrast, IL-17 facilitates S. aureus clearance by modulating local neutrophil responses.15 IL-22 and IL-17 have previously been shown to modulate distinct pathways in a keratinocyte model of psoriatic skin where IL-17 controls proinflammatory responses and neutrophil recruitment, whereas IL-22 mediates the downregulation of differentiation markers.46 In our study, IL-22 does not impact upon neutrophil responses and IL-22 does not regulate IL-17 expression. Importantly, IL-22 is also expressed at WT levels in IL-17A−/− mice following colonization (2.66±0.219 vs. 3.357±0.515 fold increase at day 7 for WT and IL-17A−/− mice, respectively). Therefore, it appears that IL-22 and IL-17 are required to work in parallel to prevent persistent colonization of the nasal cavity with S. aureus. A transgenic mouse model deficient in both IL-17 and IL-22 would be required to confirm this effect.
It will be important to establish whether IL-22 is a determinant of AMP expression and subsequent S. aureus carriage in humans. IL-22 deficiency in human acne inversa lesions correlated with reduced AMP expression in lesional tissue.47 However, it is clear from this study that IL-22 is a crucial component of the T-cell-mediated immune response elicited during S. aureus nasal colonization in vivo. IL-22 expression following colonization triggers an increase in AMP expression in order to mediate bacterial killing. Furthermore, IL-22 directly mediates downregulation of important staphylococcal-binding partners in the nose. These findings highlight the importance of T-cell immunity in controlling S. aureus colonization and propose a host response mechanism, whereby IL-22 and IL-17 facilitate distinct pathways of bacterial clearance from the nose. Further investigation into these mechanisms may provide therapeutic benefit for permanent eradication of S. aureus from the nares.
Methods
Mice. Animal experiments were conducted in accordance with guidelines of the Health Products Regulatory Authority, the competent authority in Ireland, approved by Trinity College Animal Research Ethics Committee. C57BL6/J mice were obtained from Harlan, UK. IL-22−/− mice48 were provided by our collaborator Professor Renauld. IL-17A−/− and IL-22−/− (C57BL6/J background) were bred in-house.
Bacteria. A streptomycin-resistant mutant of S. aureus strain Newman (Newman SmR)6 was grown on trypticase soy agar (TSA) at 37 °C for 18 h. Colonies were scraped from the plate, cells washed and resuspended in sterile PBS, and adjusted to 2 × 1010 CFU per ml at an optical density at 600 nm.
S. aureus nasal colonization model. Mice received sterile water containing 0.5 mg ml−1 of streptomycin 24 h before nasal inoculation and for the duration of the experiment. Mice were inoculated intra-nasally with one dose of S. aureus strain Newman SmR, 2 × 108 CFU in PBS (10 μl per nostril), or PBS alone. Alternatively, mice were left untreated or were treated with recombinant murine IL-22 (eBioscience, San Diego, CA, 200 ng per nose) in conjunction with the inoculum. On days 3, 10, 14, and 21, mice were killed. The nose was excised, homogenized in 500 μl PBS, and plated onto TSA with or without 0.5 mg ml−1 of streptomycin to quantify S. aureus CFU per nose.
For CFU enumeration in the nasopharynx, on days 3, 10, 14, and 21, the trachea was exposed and cannulated, and 500 μl of PBS was washed through the trachea to the nose. Nasopharyngeal washes were plated onto TSA containing 0.5 mg ml−1 of streptomycin to quantify CFU per ml.
For RNA extraction, noses were homogenized in 1 ml of Trizol reagent. For preparation of samples for enzyme-linked immunosorbent assay, noses were homogenized in lysis buffer (3% (w/w) NaCl, 50 nM HEPES, 1% (v/v) Igepal). Supernatant was collected and IL-22, IL17, IL-1β, IL-6, and CXCL1 concentrations were determined by enzyme-linked immunosorbent assay (R&D System, Minneapolis, MN).
RNA extraction, complementary DNA synthesis, and qPCR. RNA from homogenized tissue was extracted by Trizol reagent (Invitrogen, Waltham, MA) according to the manufacturers’ instructions. RNA yield was measured on a NanoDrop ND-1000 (Nanodrop, Wilington, DE). RNA was stored at −80 °C.
RNA (250 ng) was reverse-transcribed using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) with random primers according to the manufacturers’ instructions. mRNA was quantified using quantitative PCR on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using iTaq Sybr Green Supermix or SsoFast probes supermix (Bio-Rad) according to the manufacturers’ recommendations. Primer pairs are listed in Table 1.16, 49 Expression of mRNA from homogenized tissue was compared with that of tissue from control mice by the change-in-cycle-threshold (ΔΔCT) method.
Protein extraction from nasal tissue and western immunoblotting. Nasal tissue was excised and homogenized in 500 μl of PBS. Total protein concentration was measured using a bicinchoninic acid assay and was normalized to 500 μg ml−1. Tissue was diluted in sample buffer (Laemmli, Sigma, St. Louis, CA), heated to 95 °C for 10 min, separated on polyacrylamide gels (10–12%), and transferred onto polyvinylidene difluoride membranes. Filters were blocked in 10% (w/v) skimmed milk proteins before probing with rabbit anti-murine loricrin polyclonal IgG (Abcam, Cambridge, UK) followed by horseradish peroxidase-conjugated goat anti-rabbit IgG. Reactive bands were visualized using ECL detection system and ImageLab developing system (Bio-Rad). Bound antibody was removed using stripping buffer (Sigma) and reprobed with β-actin. Blots were stripped and reprobed with rabbit anti-murine K10 IgG followed by horseradish peroxidase-conjugated goat anti-rabbit IgG.
Flow cytometry. Nasopharyngeal tissue, including the NALT, was isolated as previously described,50 collected in sterile PBS, and filtered through a 40-μm nylon Falcon cell strainer.
Isolated NT cells were incubated with brefeldin A (5 μg ml−1) for 4 h. Cells were then resuspended in Fcγ block (1 μg ml−1) before extracellular staining with fluorochrome-conjugated antibodies against CD4 (BioLegend, San Diego, CA; clone GK1.5), CD3 (eBioscience; clone 145-2C11), CD8α (eBioscience; clone 53-6.7), TCRδ (eBioscience; clone GL3), and CD45 (eBioscience; clone 30-F11). Cells were fixed and permeablized using the Dakocytomation Intrastain Kit (Alere, San Diego, CA) before intracellular staining with fluorochrome-conjugated antibodies specific for intracellular IL-22 (eBioscience; clone 1H8PWSR) and IL-17 (eBioscience; clone 17B7). Analysis was performed with BD FACSCanto II using FACS DIVA and FloJo software (Treestar, Ashland, OR). Gates were set on respective Fluorescence Minus One.
For neutrophil isolation, tissue was dispensed in 3 ml of RPMI-1640 supplemented media containing collagenase (1 mg ml−1, Sigma) and DNase I (100 U ml−1, Sigma) for 90–120 min at 37 °C, and then passed through a 40-μm nylon Falcon cell strainer. Cells were resuspended in ammonium chloride potassium bicarbonate lysis buffer, washed in RPMI-1640 medium, and stained using the following fluorochrome-conjugated antibodies: CD11b (eBioscience, clone M1/70); Ly6G (BD Pharmingen, San Jose, CA, clone 1A8); and F4/80 (eBioscience, clone BM8).
Statistical analysis. Statistical analysis was performed using Prism Graphpad 5 software (Graphpad Software, La Jolla, CA) using a Mann–Whitney test, Kruskal–Wallis analysis of variance, or two-way analysis of variance. Comparisons for the groups were made using Dunns Multiple Comparisons test or Bonferroni post tests.
References
van Belkum, A. et al. Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 199, 1820–1826 (2009).
Mermel, L.A., Cartony, J.M., Covington, P., Maxey, G. & Morse, D. Methicillin-resistant Staphylococcus aureus colonization at different body sites: a prospective, quantitative analysis. J. Clin. Microbiol. 49, 1119–1121 (2011).
Wertheim, H.F. et al. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364, 703–705 (2004).
Cookson, B.D. Mupirocin resistance in staphylococci. J. Antimicrob. Chemother. 25, 497–501 (1990).
Baur, S. et al. A nasal epithelial receptor for Staphylococcus aureus WTA governs adhesion to epithelial cells and modulates nasal colonization. PLoS Pathog. 10, e1004089 (2014).
Mulcahy, M.E. et al. Nasal colonisation by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS Pathog. 8, e1003092 (2012).
Cole, A.M. et al. Determinants of Staphylococcus aureus nasal carriage. Clin. Diagn. Lab. Immunol. 8, 1064–1069 (2001).
Nakamura, M.M., McAdam, A.J., Sandora, T.J., Moreira, K.R. & Lee, G.M. Higher prevalence of pharyngeal than nasal Staphylococcus aureus carriage in pediatric intensive care units. J. Clin. Microbiol. 48, 2957–2959 (2010).
Emonts, M. et al. Host polymorphisms in interleukin 4, complement factor H, and C-reactive protein associated with nasal carriage of Staphylococcus aureus and occurrence of boils. J. Infect. Dis. 197, 1244–1253 (2008).
Nurjadi, D., Herrmann, E., Hinderberger, I. & Zanger, P. Impaired beta-defensin expression in human skin links DEFB1 promoter polymorphisms with persistent Staphylococcus aureus nasal carriage. J. Infect. Dis. 207, 666–674 (2013).
van Belkum, A. et al. The role of human innate immune factors in nasal colonization by Staphylococcus aureus. Microbes Infect. 9, 1471–1477 (2007).
Zanger, P., Nurjadi, D., Vath, B. & Kremsner, P.G. Persistent nasal carriage of Staphylococcus aureus is associated with deficient induction of human beta-defensin 3 after sterile wounding of healthy skin in vivo. Infect. Immun. 79, 2658–2662 (2011).
Verkaik, N.J. et al. Induction of antibodies by Staphylococcus aureus nasal colonization in young children. Clin. Microbiol. Infect. 16, 1312–1317 (2010).
Padoveze, M.C., de Jesus Pedro, R., Blum-Menezes, D., Bratfich, O.J. & Moretti, M.L. Staphylococcus aureus nasal colonization in HIV outpatients: persistent or transient? Am. J. Infect. Control 36, 187–191 (2008).
Archer, N.K., Harro, J.M. & Shirtliff, M.E. Clearance of Staphylococcus aureus nasal carriage is T cell dependent and mediated through interleukin-17A expression and neutrophil influx. Infect. Immun. 81, 2070–2075 (2013).
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008).
Sonnenberg, G.F., Fouser, L.A. & Artis, D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390 (2011).
Liang, S.C. et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203, 2271–2279 (2006).
Aujla, S.J. et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 14, 275–281 (2008).
Wolk, K. et al. IL-22 increases the innate immunity of tissues. Immunity 21, 241–254 (2004).
Boniface, K. et al. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J. Immunol. 174, 3695–3702 (2005).
Wolk, K. et al. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur. J. Immunol. 36, 1309–1323 (2006).
Ahlfors, H. et al. IL-22 fate reporter reveals origin and control of IL-22 production in homeostasis and infection. J. Immunol. 193, 4602–4613 (2014).
Duhen, T., Geiger, R., Jarrossay, D., Lanzavecchia, A. & Sallusto, F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 10, 857–863 (2009).
Nomura, T., Kabashima, K. & Miyachi, Y. The panoply of alphabetaT cells in the skin. J. Dermatol. Sci. 76, 3–9 (2014).
Cella, M. et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009).
Satoh-Takayama, N. et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 29, 958–970 (2008).
Van Maele, L. et al. Activation of Type 3 innate lymphoid cells and interleukin 22 secretion in the lungs during Streptococcus pneumoniae infection. J. Infect. Dis. 210, 493–503 (2014).
Simonian, P.L. et al. gammadelta T cells protect against lung fibrosis via IL-22. J. Exp. Med. 207, 2239–2253 (2010).
Mielke, L.A. et al. Retinoic acid expression associates with enhanced IL-22 production by gammadelta T cells and innate lymphoid cells and attenuation of intestinal inflammation. J. Exp. Med. 210, 1117–1124 (2013).
de Repentigny, L., Goupil, M. & Jolicoeur, P. Oropharyngeal Candidiasis in HIV Infection: Analysis of Impaired Mucosal Immune Response to Candida albicans in Mice Expressing the HIV-1 Transgene. Pathogens 4, 406–421 (2015).
Gauguet, S. et al. Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect. Immun. 83, 4003–4014 (2015).
Ivanov, I.I. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008).
Sanos, S.L. et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 10, 83–91 (2009).
Cho, M.L. et al. STAT3 and NF-kappaB signal pathway is required for IL-23-mediated IL-17 production in spontaneous arthritis animal model IL-1 receptor antagonist-deficient mice. J. Immunol. 176, 5652–5661 (2006).
Maher, B.M. et al. Nlrp-3-driven interleukin 17 production by gammadeltaT cells controls infection outcomes during Staphylococcus aureus surgical site infection. Infect. Immun. 81, 4478–4489 (2013).
O'Brien, L.M., Walsh, E.J., Massey, R.C., Peacock, S.J. & Foster, T.J. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell. Microbiol. 4, 759–770 (2002).
Zhang, Z., Clarke, T.B. & Weiser, J.N. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J. Clin. Invest. 119, 1899–1909 (2009).
Nograles, K.E. et al. IL-22-producing "T22" T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J. Allergy Clin. Immunol. 123, 1244–1252.e2 (2009).
Ishigame, H. et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 30, 108–119 (2009).
Midorikawa, K. et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect. Immun. 71, 3730–3739 (2003).
Cash, H.L., Whitham, C.V., Behrendt, C.L. & Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006).
Sestito, R. et al. STAT3-dependent effects of IL-22 in human keratinocytes are counterregulated by sirtuin 1 through a direct inhibition of STAT3 acetylation. FASEB J. 25, 916–927 (2011).
Choi, S.M. et al. Innate Stat3-mediated induction of the antimicrobial protein Reg3gamma is required for host defense against MRSA pneumonia. J. Exp. Med. 210, 551–561 (2013).
Son, E.D. et al. Staphylococcus aureus inhibits terminal differentiation of normal human keratinocytes by stimulating interleukin-6 secretion. J. Dermatol. Sci. 74, 64–71 (2014).
Nograles, K.E. et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br. J. Dermatol. 159, 1092–1102 (2008).
Wolk, K. et al. Deficiency of IL-22 contributes to a chronic inflammatory disease: pathogenetic mechanisms in acne inversa. J. Immunol. 186, 1228–1239 (2011).
Kreymborg, K. et al. IL-22 is expressed by Th17 cells in an IL-23-dependent fashion, but not required for the development of autoimmune encephalomyelitis. J. Immunol. 179, 8098–8104 (2007).
Kryczek, I. et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity 40, 772–784 (2014).
Asanuma, H. et al. Isolation and characterization of mouse nasal-associated lymphoid tissue. J. Immunol. Methods 202, 123–131 (1997).
Acknowledgements
This work was funded primarily by an Irish Research Council government of Ireland postdoctoral fellowship (GOIPD/2014/385) awarded to M.E.M. and a Science Foundation Ireland Investigator Project grant (12/IP/1532) awarded to RMM.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Additional information
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Mulcahy, M., Leech, J., Renauld, JC. et al. Interleukin-22 regulates antimicrobial peptide expression and keratinocyte differentiation to control Staphylococcus aureus colonization of the nasal mucosa. Mucosal Immunol 9, 1429–1441 (2016). https://doi.org/10.1038/mi.2016.24
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2016.24
This article is cited by
-
Chemerin Exacerbates Psoriasis by Stimulating Keratinocyte Proliferation and Cytokine Production
Current Medical Science (2023)
-
Advanced In Vitro Three-Dimensional Skin Models of Atopic Dermatitis
Tissue Engineering and Regenerative Medicine (2023)
-
CCR2 mediates increased susceptibility to post-H1N1 bacterial pneumonia by limiting dendritic cell induction of IL-17
Mucosal Immunology (2019)
-
Interleukin-22 and Its Correlation with Disease Activity in Plaque Psoriasis
Archivum Immunologiae et Therapiae Experimentalis (2019)