Abstract
Invariant natural killer T (iNKT) cells produce cytokines interleukin-4 (IL-4) and IL-13 during type-2 inflammatory responses. However, the nature in which iNKT cells acquire type-2 cytokine competency and the precise contribution of iNKT cell–derived IL-4 and IL-13 in vivo remains unclear. Using IL-13-reporter mice to fate-map cytokine–expressing cells in vivo, this study reveals that thymic iNKT cells express IL-13 early during development, and this IL-13-expressing intermediate gives rise to mature iNKT1, iNKT2, and iNKT17 subsets. IL-4 and IL-13 reporter mice also reveal that effector iNKT2 cells produce IL-4 but little IL-13 in settings of type-2 inflammation. The preferential production of IL-4 over IL-13 in iNKT2 cells results in part from their reduced GATA-3 expression. In summary, this work helps integrate current models of iNKT cell development, and further establishes non-coordinate production of IL-4 and IL-13 as the predominant pattern of type-2 cytokine expression among innate cells in vivo.
Similar content being viewed by others
Introduction
Pathology associated with allergic disease is a result of type-2 inflammation and represents a significant global health burden.1 Both allergic disease and helminth infections are associated with IgE production, innate cell recruitment to sites of inflammation, smooth muscle contractility, and mucus production.2 These hallmarks of type-2 inflammation are tightly associated with the expression of two key cytokines, interleukin-4 (IL-4) and IL-13 both of which serve distinct roles in vivo during type-2 immune responses.3, 4 IL-4 is required for IgE production, whereas IL-13 is sufficient to drive peripheral aspects of type-2 inflammatory responses. Importantly, IL-13 is considered the dominant cytokine involved in airway restriction during asthma and worm clearance during anti-helminth immunity.5, 6, 7
Although canonical T-helper type-2 (Th2) cells are known to be important producers of IL-4 and IL-13, many innate immune cells also produce these cytokines during type-2 inflammation. The precise contribution of IL-4 and IL-13 by various innate cells in settings of type-2 inflammation remains an important question in the field. Invariant natural killer T (iNKT) cells are one innate cell subset described to be important in settings of allergic inflammation. iNKT cells reside in various organs including lymph nodes, spleen, liver, bone marrow, and these cells appear to play particularly important roles in mucosal tissues such as the lung and intestine.8, 9 In the context of type-2 inflammation, iNKT cell–derived IL-4 and IL-13 have been shown to promote the development of airway inflammation, while interferon (IFN)-gamma is an important negative modulator.10, 11, 12, 13
Invariant NKT cells have been described to coordinately produce IL-4 and IL-13 during type-2 inflammation.11, 14 However, the precise contribution of iNKT cells and iNKT cell–derived cytokines in allergic lung inflammation remains an area of debate.15, 16, 17 Because of difficulties associated with detecting IL-4 and IL-13 in vivo, most studies to date have relied on ex vivo restimulation to assess iNKT cell cytokine potential. As such, differences between the true nature of in vivo cytokine production compared to what can be achieved after ex vivo restimulation may contribute to the disparities associated with these findings.
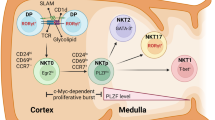
iNKT cells produce the cytokines IFN-gamma, IL-4, IL-13, and IL-17 in effector tissues suggesting that iNKT cells are able to serve various functions during an immune response.18, 19, 20, 21, 22, 23 iNKT cells acquire IL-4, IL-17, and IFN-gamma competency during development in the thymus, and ultimately mature into three distinct subsets based on transcription factor and cytokine expression.24 The subsets iNKT1, iNKT2, and iNKT17 produce IFN-gamma, IL-4, and IL-17, respectively, in a manner similar to conventional Th1, Th2, and Th17 CD4+ T-helper subsets. Like T-helper cell subsets, lineage-determining transcription factors determine the fate and commitment of iNKT cells to one of these three subsets. In the case of iNKT2 cells, GATA-3 (the Th2 lineage–determining factor) and lymphoid enhancer factor 1 are required to achieve full iNKT2 cell fate and to produce IL-4 and IL-13.24, 25
Currently, there are two models for iNKT lineage differentiation referred to as the linear differentiation model and the lineage diversification model. The linear differentiation model suggests that iNKT cells develop along three stages in the thymus, and iNKT cells first acquire transcriptional competency for IL-4, before acquiring the capacity to express IFN-gamma (and in some cases IL-17) as they undergo further maturation.18, 26, 27, 28 More recently the lineage diversification model emerged to suggest that iNKT cells producing IL-4 are distinct from those producing IFN-gamma and IL-17. This model is based on data showing that thymic iNKT cells are programmed and committed during development to express specific lineage-determining factors, and these transcription factors restrict plasticity and promote terminal fate commitment.24 In this lineage diversification model, mature iNKT cells expressing IFN-gamma, IL-4, and IL-17 arise as distinct lineages (iNKT1, iNKT2, and iNKT17, respectively). In this model, iNKT1, iNKT2, and iNKT17 subsets likely do not share a common cytokine-expressing developmental intermediate as proposed in the classical linear model of iNKT cell differentiation. Although IL-4 expression during iNKT cell development in the thymus has been studied extensively in the context of these two models, IL-13 transcriptional competency has yet to be fully characterized.18, 21, 23, 29
We used mice to lineage-trace IL-13-expressing cells and our results showed that virtually all iNKT cells found in the thymus exhibit prior IL-13 expression, a phenotype that is highly correlated with IL-4 competency. Previous IL-13 expression was evident not just in iNKT2 cells, but also committed in iNKT1 and iNKT17 subsets. These findings are consistent with a model in which mature iNKT subsets arise from a common precursor that is competent to produce both IL-4 and IL-13. In addition, models of helminth- and allergen-driven type-2 inflammation showed that lung-resident iNKT2 cells exhibited a significant preference for IL-4 production over IL-13. The differential production of IL-4 and IL-13 is likely a consequence of intermediate GATA-3 protein expression. In peripheral tissues, IL-13-producing immune cells exhibited high levels of GATA-3 expression, whereas cells with low or intermediate levels of GATA-3 preferentially produced IL-4. Thus, it is possible that GATA-3 functions as a biological rheostat for IL-4 and IL-13 production in vivo. This parallels findings observed among IL-4-restricted follicular T-helper cells and IL-4- and IL-13-producing, canonical Th2 cells.4 In summary, IL-4 and IL-13 expression is tightly regulated in iNKT cells during development and at peripheral sites of type-2 inflammation.
Results
Thymic iNKT cells express IL-13
Although IL-4 expression during iNKT cell development has been well studied, less is known with regard to IL-13 expression. In order to assess IL-13 expression directly in vivo, we analyzed IL-13 production in thymic iNKT cells using IL13YetCre13RosaYFP lineage-tracing reporter mice. IL13YetCre13RosaYFP mice generate a bicistronic transcript allowing endogenous IL-13 to be produced in conjunction with an enhanced yellow fluorescent protein (YFP)-cre recombinase fusion protein. The expression of YFP-cre fusion protein excises a transcriptional stop element in the targeted rosa26 locus inducing constitutive YFP expression.4, 30 In brief, IL13YetCre13RosaYFP mice permanently label all cells that produced IL-13 mRNA with YFP for the lifespan of the cell.
Approximately 95% of CD44high, thymic iNKT cells found in naive mice exhibited evidence of current or prior IL-13 expression (Figure 1a,b). Both CD4+ and CD4− iNKT cells expressed YFP equally (Figure 1a,b; Supplementary Figure 1 online). We observed the same findings among iNKT cells isolated from the thymus of mice exposed to Nippostrongylus brasiliensis, a helminth that induces a robust type-2 inflammatory response (Supplementary Figure 1 & 2). Of note, the near ubiquitous expression of IL-13 observed among thymic iNKT cells was unique to this T-cell subset as conventional T cells in the thymus remained largely negative for YFP expression throughout development (Figure 1b; Supplementary Figure 1 & 2d). These findings also held true in the periphery as virtually all iNKT cells analyzed from the spleen, lung, and liver of naive mice showed evidence of prior IL-13 expression (Figure 1c), while only 1–2% of CD4+ T cells showed evidence of current or prior IL-13 expression in these same tissues.
Invariant natural killer T (iNKT) cells are programmed for interleukin-4 (IL-4) and IL-13 competency during development. (a) Representative flow cytometry plots for yellow fluorescent protein (YFP) expression in thymic CD4+ invariant NKT (iNKT) and CD4− iNKT cells harvested from naive IL13YetCre13/+RosaYFP/+ mice. (b) Percentage of YFP expression among naive thymic iNKT cells (TCRβ+PBS57+), CD4+ (CD4+PBS57−), CD8+ (CD8+PBS57−), double-positive (CD4+CD8+PBS57−), and double-negative (CD4−CD8−PBS57−) conventional T cells from IL13YetCre13/+RosaYFP mice. (c) Percentage of YFP+ CD4+ TCRβ+ T cells (open circles) and iNKT cells (closed circles) in indicated organs of naive mice. Data are representative of two independent experiments with two to five mice per group.
In an effort to compare how these results relate to IL-4 expression, we turned to IL44get reporter mice, where cells actively generating IL-4 mRNA are marked by green fluorescent protein (GFP) expression.31 We observed that the proportion of iNKT cells that expressed GFP was reduced compared to the proportion of iNKT cells fate-mapped for IL-13 (Supplementary Figure 2). This difference indicates either that IL-13 and IL-4 are not coordinately expressed during iNKT cell development or that a subset(s) of thymic iNKT cells loses the ability to constitutively express type-2 cytokines. The latter would be expected in the case of iNKT1 and iNKT17 cell generation.24
iNKT cell subsets share a common IL-13-expressing intermediate
The observed difference between IL-4 and IL-13 reporter expression among thymic iNKT cells led us to further characterize IL-13-fate-mapped iNKT cells. As iNKT2 cells make up 20% of the total thymic iNKT cells in 6- to 8-week-old C57BL6 mice, the data were most consistent with iNKT cells passing through an early, IL-13-expressing intermediate prior to their eventual commitment to iNKT1, iNKT2, or iNKT17 cell fates.24 Expression of IL-13 within an uncommitted iNKT cell intermediate is consistent with prior studies showing that iNKT cells express IL-4 early during development.18, 27, 29 By using the classical linear differentiation model where iNKT cells develop from CD44low, NK1.1− (stage 1) to CD44high, NK1.1− (stage 2), to CD44high, NK1.1+ (stage 3), prior publications have placed initial IL-4 expression between stages 1 and 2.29 To test if IL-13 expression occurs at a similar developmental point, YFP expression was assessed at different developmental stages in thymic CD1d–positive iNKT cells isolated from naive IL13YetCre13RosaYFP mice (Figure 2a,b). Similar to previous findings for IL-4, IL-13 is initially expressed late in stage 1 and early in stage 2 (Figure 2b,c). Again, IL-13 expression in thymic iNKT cells did not show a bias toward CD4 expression as both CD4+ and CD4− populations are fate-mapped equivalently in stages 2 and 3 (Figure 2c). Expression of IL-13 in CD4+ and CD4− thymic iNKT subsets is consistent with previous reports.21
Thymic invariant natural killer T (iNKT) cell subsets pass through an interleukin-13 (IL-13)–expressing stage early during development. (a) Representative flow cytometry plots of thymic iNKT cells harvested from naive IL13YetCre13/+RosaYFP/+ or IL13YetCre13/+mice. (b) Percentage of yellow fluorescent protein (YFP) expression among developing thymic iNKT cells CD44low, NK1.1low (stage 1); CD44high, NK1.1low (stage 2); and CD44high, NK1.1 positive (stage 3) iNKT T cells from IL13YetCre13/+mice (top panel) IL13YetCre13/+RosaYFP mice (bottom panel). (c) Percentage of YFP+ iNKT cells at indicated stages of development. Data are cumulative of three independent experiments; four mice total per group. (d) Total iNKT cells from IL13YetCre13/+ mice or YFP-expressing iNKT cells from IL13YetCre13/+RosaYFP/+ were sorted and stained for intracellular transcription factors. Gates depict indicated iNKT cell subsets. Data are representative of three independent experiments (one to two mice per group).
The above findings suggest that iNKT cells pass through a common developmental stage in which IL-13 is expressed prior to final commitment of iNKT1, iNKT2, and iNKT17 cells. To further test if iNKT1, iNKT2, and iNKT17 cells pass through an IL-13 intermediate, we sorted YFP+ (IL-13 fate-mapped) iNKT cells from the thymus of IL13YetCre13RosaYFP mice and stained for lineage-determining factors that distinguish iNKT1, iNKT2, and iNKT17 cells. Using total iNKT cells from IL13Yetcre13 mice as a control, we were able to distinguish iNKT1 (PLZFlow, T-bethigh) and iNKT2/iNKT17 (PLZFhigh/Int, GATA-3high) subsets as previously described (Figure 2d).24, 25 IL-13 fate-mapped iNKT cells showed the same pattern of staining as total iNKT cells indicating that iNKT1, iNKT2, and iNKT17 cells express IL-13 at some point upstream of their final fate commitment. When and how iNKT1 and iNKT17 cells turn off IL-13 expression remains an intriguing question for future study.
iNKT cells represent a small fraction of the total IL-4-competent cells during helminth exposure
To better understand the contribution of iNKT cells in type-2 inflammation, we sought to obtain a comprehensive view of the cell types capable of producing IL-4 and IL-13 to assess the overall impact of cytokine production by iNKT cells. To induce type-2 inflammation, we exposed IL44get/4get mice to N. brasiliensis and accounted for all IL-4-competent cells found in the lung during the course of infection. Although a small number of GFP-expressing cells were found in the lung of uninfected mice, the percentage of GFP-expressing cells increased significantly over the course of infection (Figure 3a). Sixty percent of live cells in the lung were competent for IL-4 expression within 8 days of helminth exposure. This represented a 122-fold increase in the total number of IL-4-competent cells over that found in uninfected mice (Figure 3b). We identified GFP+ cells as CD4+ T cells (T cell receptor-β-positive), iNKT cells, small non-B non-T (NBNT) cells, basophils, and eosinophils (Figure 3c). CD4+ T cells, basophils, and eosinophils represented the majority of IL-4-competent cells at all time points tested (Figure 3d).
Quantification of interleukin-4 (IL-4)-competent cells after infection with Nippostrongylus brasiliensis. (a) Flow cytometry analysis of green fluorescent protein (GFP; IL-4) expression in the lung of IL44get/4get mice on days 0, 2, 5, and 8 post infection with N. brasiliensis. Gates in contour plots and bar graph represent the percentage of GFP+ cells (mean±s.e.m.). (b) Enumeration of total GFP-positive cells in the lung of IL44get/4get mice on days 0, 2, 5, and 8 post infection with N. brasiliensis. Summary of results (mean±s.e.m.) (c) Representative gating strategies for CD4+ T cells (blue), natural killer T (NKT) cells (green), non-B non-T (NBNT, gray), basophils (red), and eosinophils (black), in the lung of IL44get/4get mice 8 days after infection with N. brasiliensis. Gates represent the percentage of cells among the indicated population stated above the plot. (d) Enumeration of viable, GFP-positive TCRβ+CD4+ T cells (blue), NKT cells (green), NBNT (gray), basophils (red), and eosinophils (black) in the lung of IL44get/4get mice on 0, 2, 5, and 8 days post infection (dpi) with N. brasiliensis (mean±s.e.m.). Data are representative of at least two independent experiments with three to five mice per experimental group. (e) Flow cytometry plots of lung invariant NKT (iNKT) cells depicting CD4 and GFP (IL-4 mRNA) expression in IL44get/4get mice at indicated time points post N. brasiliensis exposure. (f) Graph depicts the percentage of CD4+ (closed circles) and CD4− (open circles) iNKT cells expressing GFP (IL-4) among total iNKT cells (mean±s.d.). Data are cumulative of two independent experiments.
GFP+, CD1d-restricted iNKT cells and NBNT cells also increased throughout infection, but these cells remained a minor fraction of the total IL-4-competent population in the lung (Figure 3d). Among iNKT cells in the lung during the course of infection, IL-4 competency is enriched among CD4+ iNKT cells consistent with prior findings that identified CD4 as a marker for iNKT2 cells and a subset of NKT1 cells (Figure 3e,f).14, 24, 25 In summary, these results show that while iNKT cells can be readily detected in the lung of both naive and helminth infected mice, they represent a small fraction of the total IL-4-competent pool in vivo.
iNKT cells are an early contributor of IL-4 but not IL-13 protein in settings of type-2 inflammation
The presence of IL-4 mRNA does not always correlate with protein production in vivo.32 As such, we sought to quantify the amount of cytokine protein produced by iNKT cells in an inflamed lung. To do this, we used additional cytokine reporter mouse lines to assess IL-4 and IL-13 protein production. IL4KN2 mice report IL-4 protein by placing a human CD2 marker into the endogenous il4 locus (Figure 4a). Human CD2 labels cells that have recently produced IL-4 protein.32 Similarly, IL13 Smart13 mice report IL-13 production by inserting a human CD4 marker at the endogenous IL-13 locus (Figure 4a). These mice faithfully report IL-13 protein production in vivo.4 IL4KN2 and IL13Smart13 reporter lines were crossed to IL44get mice to restrict the analysis of IL-4 and IL-13 protein production to cells competent for type-2 cytokine expression after helminth exposure. These dual reporter mice allow the direct assessment of IL-4 and IL-13 protein production in vivo during helminth infection (Figure 4b).
Examination of the cellular sources of biologically active interleukin-4 (IL-4) and IL-13 during type-2 inflammation. (a) Description of IL44get, IL4KN2, and IL13Smart13 mice. (b) Experimental design—Nippostrongylus brasiliensis infection and analysis of IL4KN2/4get or IL44get/+IL13Smart13/+ mice. (c) Percentage of huCD2+ staining within viable, green fluorescent protein (GFP)–positive CD4+ T cells, invariant natural killer T (iNKT) cells, group 2 innate lymphoid (ILC2), basophils, and eosinophils in the lung of IL4KN2/4get mice 8 days (closed bars) after infection with N. brasiliensis or left uninfected (open bars; mean±s.d.). (d) Absolute number of viable huCD2+GFP+ cells in the lung of IL4KN2/4get mice 8 days after N. brasiliensis infection (mean±s.e.m.). (e) Percentage of huCD4+ staining within viable, GFP-positive CD4+ T cells, NKT cells, ILC2, basophils, and eosinophils in the lung of IL44get/+IL13Smart13/+ mice 8 days (closed bars) after infection with N. brasiliensis or left uninfected (open bar; mean±s.d.). (f) Absolute number of viable huCD4+GFP+ cells in the lung of IL44get/+IL13Smart13/+ mice 8 days after N. brasiliensis infection (mean±s.e.m.). Data are representative of at least two independent experiments with three to five mice per experimental group.
Analysis of IL4KN2/4get mice 8 days after N. brasiliensis exposure revealed that only 15–20% of GFP+ CD4+ T cells produced IL-4 protein in the lung as indicated by human CD2 expression (Figure 4c). This was significantly higher than what was observed on day 5 of infection where <5% of lung CD4+ T cells showed evidence of IL-4 protein production (Supplementary Figure 3C). Thus, although CD4+ T cells are transcriptionally competent for IL-4 (GFP+), protein production (human CD2) remained a small subset within the GFP+ population in vivo. This restricted cytokine production was also observed in GFP+ iNKT cells and GFP+ basophils (Figure 4c; Supplementary Figure 3C), in which 10–20% of cells expressing IL-4 mRNA (GFP+) showed evidence of IL-4 protein production (human CD2) 5 and 8 days after helminth exposure. Less than 2% of GFP+ eosinophils and group 2 innate lymphoid (ILC2) cells produced IL-4 protein at these time points (Figure 4c; Supplementary Figure 3C).
Interestingly, total cell counts revealed that CD4+ T cells, eosinophils, and basophils were the major contributors of IL-4 protein 5 and 8 days after helminth infection (Figure 4d; Supplementary Figure 3D). In contrast, although a high percentage of the iNKT cell population produced IL-4, the small number of iNKT cells present in the lung minimized their contribution to the total IL-4-producing pool (Figure 4c,d; Supplementary Figure 3C,D).
We next performed an analysis of IL-13 production in the lung of helminth-exposed mice (Figure 4a,b). Assessment of IL44get/+IL13Smart13/+ mice showed that, similar to IL-4, IL-13 protein production was restricted among IL-4-competent populations (Figure 4e). Approximately 15% of IL-4-competent (GFP+), CD4+ T cells and nearly 40% of GFP+ ILC2 cells produced IL-13 at the peak of the response to N. brasiliensis (Figure 4e). Although the percentages were overall lower, the same trend held true at day 5 of infection (Supplementary Figure 3E). In contrast to what was observed for IL-4 production, a very low percentage of iNKT cells and basophils produced IL-13 protein (Figure 4e).
Similar to IL-4 production, total cell counts revealed that CD4+ T cells were the predominant IL-13-producing population in the infected lung at both day 5 and day 8 of the response (Figure 4f; Supplementary Figure 3F). Despite the high percentage of IL-4-competent ILC2 cells producing IL-13, these cells contribute a relatively small amount of IL-13 compared to CD4+ T cells, and again, the small number of iNKT cells in the inflamed lung minimized their overall contribution to the IL-13-producing pool (Figure 4f; Supplementary Figure 3F).
Restricted IL-13 protein expression was recapitulated in a second model of type-2 lung inflammation using a house dust mite (HDM) Dermatophagoides pteronyssinus–induced model of allergic airway inflammation (Figure 5a).33 In this case, IL44get/+IL13Smart13/+ mice were sensitized and challenged with HDM, and IL-13 production was assessed in the lung 2 days after the last challenge. Similar to what was observed during N. brasillensis infection, CD4+ T cells were the predominant producer of IL-13, while iNKT cells and other innate cells contributed minimally to the total IL-13-producing pool (Figure 5b,c). Unlike CD4+ T cells, which showed a significant increase in IL-13 production after challenge, iNKT cells showed no such increase (Supplementary Figure 4C–F). These findings indicated that while iNKT cells producing IL-4 and IL-13 protein are detectable, they are the source of only a small fraction of the total IL-4 and IL-13 in the lung. This result is intriguing given that iNKT-derived IL-13 has been suggested to play a substantial role in airway restriction after allergen challenge in other models of allergic lung inflammation.10, 11 With regard to helminth infection and the HDM model of type-2 inflammation used here, the data suggest that IL-4-producing iNKT cells, rather than IL-13-producing iNKT cells, play an important role early during a typical type-2 response. However, the data also suggest that these cells likely have a reduced impact once type-2 inflammation is firmly established. The reporter mice described herein should prove helpful in understanding the differential requirement for iNKT cell–derived cytokines in different model systems of allergic lung inflammation.
CD4+ T cells are the primary source of interleukin-13 (IL-13) during house dust mite (HDM)–induced allergic inflammation. (a) Experimental design—HDM model of allergic inflammation. (b) Percentage of huCD4+ staining within viable green fluorescent protein (GFP)-positive CD4+ T cells, invariant natural killer T (iNKT) cells, group 2 innate lymphoid (ILC2), basophils, and eosinophils in the lung of IL44get/+IL13Smart13/+ mice after sensitization (intraperitoneal) and challenge (intranasal) with HDM. (c) Absolute number of viable huCD4+GFP+ cells in the lung of IL44get/+IL13Smart13/+ mice after sensitization and challenge with HDM (mean±s.e.m.). Data are representative of two independent experiments with three to five mice per experimental group.
Innate immune cells preferentially express either IL-4 or IL-13 during type-2 inflammation
The differences observed in IL-4 and IL-13 production within innate immune cells in response to N. brasiliensis suggested that these cytokines are often differentially expressed in vivo. To more thoroughly test this, IL4KN2/4get and IL44get/+IL13Smart13/+ mice were exposed to N. brasiliensis and IL-4 and IL-13-reporter expression was assessed on day 8 post infection. Although CD4+ T cells in the lung produced IL-4 and IL-13 at nearly equivalent levels, innate immune cells largely produced either IL-4 or IL-13 (Figure 6a,b). Specifically, basophils, eosinophils, and iNKT cells showed a strong preference for IL-4 production. In contrast, ILC2 cells exhibited a strong preference for IL-13 production in our analyses (Figure 6b). We also performed this analysis using the IL4KN2/+IL13Smart13/+ dual reporter system to allow direct examination of IL-4 (huCD2) and IL-13 (huCD4) protein expression within the same cell (Supplementary Figure 5A,B). The results from these experiments mirrored observations made at day 8 of helminth exposure (Figure 6a,b).
Innate immune cells display a preference for interleukin-4 (IL-4) or IL-13 during allergic inflammation. (a) Gate represents frequency of huCD4+ expression within green fluorescent protein-positive (GFP+) cells of the indicated cell type harvested from the lung of IL44get/+IL13Smart13/+ mice 8 days after infection with Nippostrongylus brasiliensis (top row). Gate represents the frequency of huCD2+ expression within GFP+ cells of the indicated cell type harvested from the lung of IL4KN2/4get mice (bottom row). (b) Ratio of the percentage of IL-13-producing cells to IL-4-producing cells within the indicated cell type 8 days after infection with N. brasiliensis. Line represents the mean ratio within each population. Data are representative of at least two independent experiments with three to five mice per experimental group.
To assess whether the preferential cytokine production among innate immune cells also held true early in response to N. brasiliensis, we assessed IL-4 and IL-13 production in IL4KN2/4get and IL44get/+IL13Smart13/+ mice 5 days after helminth exposure. Although ILC2 cells and eosinophils exhibited a less dramatic difference than that observed at day 8, non-coordinate production of these type-2 cytokines was still observed (Supplementary Figure 5C,D). Importantly, the preference for IL-4 production over IL-13 by iNKT cells was more pronounced early in the response again suggesting that cytokines from iNKT cells may play a more important role in the initial stages of type-2 inflammation. In summary, these findings challenge the paradigm of coordinate expression of type-2 cytokines as it pertains to innate cells during type-2 inflammation.
IL-13-producing innate cells require high GATA-3 expression
Previous studies suggested that GATA-3 expression is an important factor for in vivo IL-13 production.4 Thus, with the observation that iNKT cells induced during type-2 inflammation did not produce IL-13 in vivo, we hypothesized that GATA-3 expression was likely low in NKT2 cells relative to other IL-13-producing cells in the lung. To test this, mice were exposed to N. brasiliensis and lung-resident immune cells were stained for intracellular GATA-3 protein 8 days post infection (Figure 7a). CD4+ T cells were used to normalize the GATA-3high compartment. As observed previously, CD90+, Iducible T cell costimulator (ICOS)+ NBNT cells contained a GATA-3high population similar to that achieved by canonical Th2 CD4+ T cells (Figure 7a).4 Basophils, which do not express IL-13, exhibited little or no expression of GATA-3 (Figures 6a and 7a). Like basophils, very few iNKT cells fell within the range of high GATA-3 expression (Figure 7a). Our data suggest that basophils and iNKT cells are unable to maintain high GATA-3 expression, and therefore do not readily produce IL-13. Thus, we hypothesized that a threshold of GATA-3 expression was required to achieve optimal IL-13 production. To further investigate this hypothesis, we sorted CD4+ T cells, ILC2 cells, basophils, and iNKT cells from IL44get mice infected with N. brasiliensis to assess GATA-3 protein by immunocytochemistry (Figure 7b). CD4+ T cells and ILC2 cells, which produce IL-13 in the lung, were largely replete with GATA-3 as indicated by robust staining (Figure 7b). In contrast, GATA-3 staining was greatly reduced in basophils and iNKT cells, which do not produce IL-13 under these conditions in vivo.
High GATA-3 expression discriminates cells capable of producing interleukin-13 (IL-13) protein in the inflamed lung. (a) GATA-3 expression as determined by flow cytometry in indicated cell types harvested from the lungs of mice 8 days after infection with Nippostrongylus brasiliensis. Number in histogram plot and graph represents the percentage of cells present in the GATA-3 high gate. The GATA-3 high gate in each histogram plot was set relative to GATA-3 high expression in CD4+ T cells for each staining cohort. Values represent the mean±s.e.m. (b) GATA-3 expression as determined by fluorescence microscopy in viable, green fluorescent protein (GFP)–positive CD4+ T cells, group 2 innate lymphoid (ILC2), basophils, and invariant natural killer T (iNKT) cells sorted from IL44get mice 8 days after infection with N. brasiliensis. Data are representative of at least two independent experiments with three to five mice per experimental group.
Ex vivo restimulation is sufficient to artificially drive IL-13 production in iNKT cells
IL-13 is critical for many hallmarks of type-2 inflammation, however, whether iNKT cell–derived IL-13 is necessary to drive the pathology associated with type-2 inflammation remains controversial.15 We believe much of the conflicting data surrounding IL-13 from iNKT cells may stem from the use of ex vivo restimulation to assess iNKT function. In essence, ex vivo restimulation can induce cytokine production in any cell containing accessible cytokine loci. Consequently, restimulation may not accurately reflect the true nature of cytokine production in vivo. As proof of principle, we sorted GFP+ immune cells from the lung of N. brasiliensis–infected IL44get/+IL13Smart13/+ mice, and cultured them with or without phorbyl 12-myristate 13-acetate/ionomycin for 5 h. CD4+ T cells showed a threefold increase in IL-13 production after restimulation (Figure 8a). Strikingly, basophils exhibited a fivefold increase in IL-13 production after restimulation (Figure 8a). However, not all cells responded alike to restimulation, as eosinophils expressed no IL-13 protein even after treatment (Figure 8a). Thus, ex vivo treatment with phorbyl 12-myristate 13-acetate and ionomycin has differential effects on individual subsets of immune cells. Important to this discussion, restimulation has the potential to induce IL-13 expression from cells that rarely secrete IL-13 in vivo, as was shown here for basophils.
Intermediate expression of GATA-3 in invariant natural killer T (iNKT) cells is regulated by proteasomal degradation. (a) Representative contour plots of viable, green fluorescent protein (GFP)–positive CD4+ T cells, eosinophils, and basophils sorted from the lung of IL44get/+IL13Smart13/+ mice infected with Nippostrongylus brasiliensis after 5-h in vitro stimulation with or without phorbyl 12-myristate 13-acetate (PMA) and ionomycin. Gate represents the percentage of huCD4+ cells within each plot. (b,c) Percentage of huCD4+ (interleukin-13 (IL-13)), b) and mean fluorescence intensity (MFI) of GATA-3 expression (c) by GFP+ NKT cells sorted from the liver of naive IL44get/+IL13Smart13/+ mice after 5 h of in vitro PMA and ionomycin stimulation. Gates (b) and bars (c) represent the huCD4-negative (gray), huCD4-intermediate (black), and huCD4-high, foward scatter (FSC)-low expressing NKT cells (red). (d) Histogram plot of intracellular GATA-3 staining in CD4+ T cells (black), NKT cells (red), and B220+/CD11b+ cells (gray; filled) harvested from the lung of IL44get/+IL13Smart13/+ mice 8 days after infection with N. brasiliensis. Graph represents the MFI of GATA-3 in indicated populations (mean±s.e.m.). (e) Graph represents RT–PCR analysis of the relative expression GATA-3 mRNA in viable, GFP-positive CD4+ T cells, eosinophils, NKT cells, and basophils sorted from the lung of IL44get/+IL13Smart13/+ mice 8 days after infection with N. brasiliensis (mean±s.e.m.). (f) Histogram plots and graph showing GATA-3 expression after intracellular staining of in vitro-cultured GFP+ NKT cells sorted from the spleen of naive IL44get/4get mice cultured in IL-2 with (black; open histogram) or without (gray; closed histogram) 10 μM MG132 for 12 h. Statistical significance was determined by two-tailed unpaired t-tests where *P<0.05, **P<0.005, and ***P<0.0001. Data are representative of at least two independent experiments.
When GFP+ iNKT cells from the liver of IL44get/+IL13Smart13/+ mice were restimulated, we observed a 50-fold increase in huCD4 (IL-13) expression (Figure 8b). Induced IL-13 production in restimulated iNKT cells correlated with increased GATA-3 expression (Figure 8c). iNKT cells that expressed the highest levels of IL-13 protein contained roughly twofold more GATA-3 than cells that did not express cytokine after restimulation (Figure 8c). Thus, the ability of a cell to rapidly produce cytokine after ex vivo restimulation reflects its cytokine production potential, however, it may not accurately reflect actual cytokine production in vivo.
The preferential production of IL-4 over IL-13 by iNKT cells and their decreased GATA-3 expression in settings of type-2 inflammation suggested that GATA-3 might be regulated differently in these cells. To address this question, we investigated GATA-3 expression in lung-resident iNKT cells. Lung-resident B220 or CD11b-expressing cells served as negative controls, as these populations do not express detectable levels of GATA-3. We observed that GATA-3 levels in iNKT cells fell between levels found in GATA-3high and GATA-3low populations of CD4+ T cells in the lung (Figure 8d). Importantly, these findings show a stratification of GATA-3 levels among GATA-3-expressing populations. Given that GATA-3intermediate iNKT cells and GATA-3low CD4+ T cells do not produce IL-13, we hypothesized that both low and intermediate levels of GATA-3 expression are insufficient to initiate or maintain IL-13 production in vivo.
Inhibition of proteasome function increases GATA-3 accumulation in iNKT cells
Intermediate expression of GATA-3 appeared to be a characteristic unique to iNKT cells compared to other IL-4- and IL-13-producing populations. To understand how GATA-3 is maintained at an intermediate level in iNKT cells, we assessed relative levels of GATA-3 mRNA ex vivo. To do this, we infected IL44get/+IL13Smart13/+ mice with N. brasiliensis and sorted GFP-expressing CD4+ T cells, eosinophils, basophils, and iNKT cells from the lung 8 days after helminth exposure. As expected, GATA-3 mRNA was highly expressed in GFP+ CD4+ T cells (Figure 8e). Also consistent with our previous findings, basophils and eosinophils, which largely do not produce IL-13, exhibited undetectable levels of GATA-3 mRNA (Figure 8e). GATA-3 mRNA, like GATA-3 protein, in iNKT cells was intermediate to that expressed by GFP+, CD4+ T cells (Figure 8e). This confirms that gata3 transcript is present but reduced in iNKT cells compared to IL-13-expressing cell types in the lung.
We hypothesized that levels of GATA-3 protein were regulated by active proteasomal degradation. Degradation would limit GATA-3-mediated transcription of IL-13 as well as GATA-3-mediated auto-activation.34 Thus, this mechanism could account for both reduced GATA-3 protein and mRNA in iNKT cells. To test this possibility, we isolated GFP+ iNKT cells from the spleen, and cultured them in the presence or absence of the proteasome inhibitor MG132. In support of proteasomal degradation being a key regulator of GATA-3 protein accumulation in iNKT cells, we observed a significant increase in GATA-3 protein after 12 h of culture in the presence of MG132 (Figure 8f). Thus, one potential mechanism by which iNKT cells could maintain intermediate GATA-3 levels is by reducing transcription at the gata3 locus. This may be accomplished, at least in part, by degrading GATA-3 protein and effectively limiting its availability for auto-activation. Certainly, ubiquitinases represent likely mediators in this process as these proteins are known to play an important role in type-2 inflammation and can influence GATA-3 protein stability.35, 36, 37, 38
Discussion
These studies have revealed important insights with respect to iNKT cell development in the thymus. Until recently, it was believed that thymic iNKT cells progressed through a linear differentiation program. This model inferred that iNKT cells first achieved IL-4 competency and later gained the ability to make IFN-gamma.26, 27, 28 However, the linear model has been challenged recently by elegant work showing that iNKT cells producing IL-4 (referred to as iNKT2 cells) represent a distinct subset from those producing IFN-gamma (iNKT1 cells) and IL-17 (iNKT17).24 Important to this conclusion, the authors showed that thymic iNKT2 cells producing IL-4 protein were unable to give rise to iNKT1 cells. This finding was interpreted to suggest that iNKT1 and iNKT17 cells arise independently from an iNKT2 cell intermediate and therefore would likely not share a common, type-2 cytokine-competent precursor. Thus, a lineage-determining model, similar to that used by CD4+ T-helper cells, was proposed as an alternative to the classic linear developmental pathway.
To investigate these models and more specifically IL-13 expression during iNKT cell development, we used lineage tracing to track the fate of all cells that had previously expressed IL-13. Unexpectedly, these studies showed that virtually all iNKT cells in the thymus, lung, liver, and spleen of naive mice exhibited previous IL-13 expression, a result that is highly correlative with IL-4 mRNA expression. Furthermore, these IL-13 fate-mapped iNKT cells consisted of iNKT1, iNKT2, and iNKT17 subsets. This result indicates that committed iNKT1, iNKT2, and iNKT17 cells share a common intermediate that is competent for IL-4 and IL-13 expression. The data provided are consistent with this intermediate arising late in stage 1 of the classical linear differentiation pathway. One potential mechanism to account for why iNKT1 and iNKT17 cells lose the ability to generate IL-13 may be through the orchestrated expression of lineage-determining transcription factors such as T-box expressed in T cells (T-bet) and RORγt starting in stage 2. Expression of T-bet or RORγt would promote the terminal differentiation of iNKT1 and iNKT17 subsets while at the same time restricting iNKT2 commitment and IL-13 production by blocking GATA-3 expression. In support of varying levels of GATA-3 affecting cell-fate determination, analysis of ILC cell subsets show that subtle changes in GATA-3 levels play a major role in ILC2 (IL-13-expressing) vs. ILC3 (IL-17-expressing) cell commitment.39 Thus, it is interesting to speculate that iNKT cell subset commitment during development may be regulated in a similar manner. In summary, the identification of a common IL-4/IL-13-competent precursor helps to unify components of the linear and lineage diversification models into a general model of iNKT cell development and lays the foundation to explore the role of GATA-3 and other lineage-determining factors in iNKT cell–fate decisions.
The present study also indicates that cytokine regulation by iNKT cells in tissues undergoing robust type-2 inflammation is more restricted than previously appreciated. The prevailing theory argues that IL-4 and IL-13 are co-expressed by immune cells during type-2 inflammation. This largely stems from work assessing cytokine production in Th2-cell clones or after in vitro restimulation of innate immune cells.40, 41, 42, 43 Indeed, direct in vivo assessment confirmed that canonical Th2 cells largely exhibit coordinate regulation of IL-4 and IL-13, but innate immune cells, such as basophils and ILC2 cells, showed a distinct preference for only IL-4 or IL-13, respectively.4 In this study, we have extended these observations to show that iNKT cells also exhibit non-coordinate regulation of IL-4 and IL-13, with a strong preference for IL-4 production.
Regulation of GATA-3 expression and protein production is one potential mechanism to explain the differential type-2 cytokine production observed in iNKT cells. Although previous studies in Th2 cells have shown that the expression of IL-5 and IL-13 have greater dependency on GATA-3 than IL-4, whether this was true among innate immune cells remained unclear.44, 45, 46 Evidence that GATA-3 is indeed essential for IL-13 production by innate cells comes from studies showing that GATA-3 expression is critical for ILC2 development, function, and IL-13 production.4, 47, 48 Although GATA-3 expression in iNKT cells was significantly lower than that found in ILC2 and Th2 cells, consistent with previous findings, the amount of GATA-3 was significantly higher than that found in naive CD4+ T cells.49 Thus, it is likely that a threshold of GATA-3 is needed in order to enter the nucleus in sufficient quantities to promote both GATA-3-mediated auto-activation and transcription of il13.34, 50 High amounts of GATA-3 protein would also promote epigenetic changes at the il13 locus. Such epigenetic changes are critical in order to establish and maintain IL-13 production in vivo.51 Thus, cells that do not maintain GATA-3 protein above this critical threshold will preferentially express IL-4 instead of IL-13. In support of such a model, previous work has shown that increased intracellular GATA-3 levels resulted in amplified IL-13 production by ILC2 cells as compared to animals lacking the transgene.52 Furthermore, a twofold reduction in GATA-3 was shown to completely block IL-13 production in ILC2 cells after restimulation. Taken together, these prior studies in ILC2 cells and those presented here for iNKT cells are consistent with a cell needing to reach a specific threshold of GATA-3 protein in order to achieve the ability to prominently produce IL-13 in vivo.4, 52 Another parallel between ILC2 and iNKT2 cells is their ability to respond to the alarmin IL-25 and produce IL-13.14, 23, 53, 54 Whether IL-25 plays a significant role in the production of IL-13 in iNKT2 cells remains an intriguing possibility.
To conclude, in this study we have identified that the differential expression of IL-4 and IL-13 among various innate immune cells likely plays an important role in diseases characterized by type-2 inflammatory responses. As IL-13 drives many of the detrimental hallmarks associated with type-2 inflammation, treatments designed to ablate IL-13-producing subsets or inhibit cytokine production specifically in these populations could be used to ameliorate allergic inflammation. Blocking IL-13 independently of IL-4 has shown promise in clinical trials, and the studies described herein provide a mechanistic framework in which to further test such targeted therapies.55 Furthermore, these studies highlight the potential for specifically targeting GATA-3 for proteasomal degradation in order to reduce IL-13 expression in immune cells. A greater understanding of such pathways will likely uncover novel therapeutic options to limit IL-13-mediated allergic pathology.
Methods
Mice
IL44get, IL4KN2, IL13Smart13, IL13YetCre13, and RosaYFP mice have been previously described.4, 30, 31 For this study, IL44get, IL4KN2/4get, and IL44get/+IL13Smart13/+ mice were bred onto the inbred Balb/c genetic background. The IL13YetCre13 and RosaYFP mice were maintained on the inbred C57BL/6 genetic background. Mice were maintained in specific pathogen-free animal facilities in accordance with animal guidelines established by the Division of Laboratory Animal Resources (DLAR), Institutional Animal Care and Use Committee (IACUC), and Duke University Medical Center (DUMC).
Infection
N. brasiliensis was prepared as previously described.56 Mice were infected with 500 L3 larvae in 200 μl phosphate-buffered saline (PBS) subcutaneously in the lower back.
HDM model of asthma
Asthma was induced as previously described.33 Briefly, mice were sensitized by intraperitoneal injection with 50 μg D. pteronyssinus in PBS (Greer Laboratories, Lenoir, NC) on days 1, 3, 5, 7, 9, 11, and 13. Mice were challenged (intranasal) with 50 μg D. pteronyssinus in PBS on days 25, 26, and 27. Mice were killed on day 29 for analysis.
Flow cytometry and cell counting
Lungs from naive and infected mice were harvested on the indicated day and single-cell suspensions were prepared. Cells were stained with the following antibodies: CD4 (BV605 or PerCP/Cy5.5), B220 (PerCP/Cy5.5), CD11b (PerCP/Cy5.5 or BV605), CD8 (PerCP/Cy5.5), CD3 (PerCP/Cy5.5), GR1 (PerCP/Cy5.5), TCRβ (APC780), CD90 (APC/Cy7), ICOS (PE or APC), DX5 (PE/Cy7), Siglec-F (PE), CD131 (PE), huCD2 (PE or APC), huCD4 (PE or APC), GATA-3 (FITC or PE or PE/Cy7 or eFluor660), T-bet (PE-Cy7), and PLZF (PE). NKT cells were identified by positive staining with α-galceramide-loaded CD1d tetramer (PBS57; provided by the National Institute of Health tetramer core). Lineage negative gate for ILC2 (CD90+, ICOS+) cell determination consisted as either CD4−, CD8−, TCRb−, CD11b−, and B220− or CD4−, CD8−, CD3−, CD11b−, Ly6C/G−, and B220−. Intracellular staining for lineage-determining transcription factors PLZF, T-bet, and GATA-3 was performed using Foxp3-staining buffer set (eBioscience, San Diego, CA) according to manufacturer’s instructions. Viable cells were identified as 4′,6-diamidino-2-phenylindole, dihydrochloride negative. Total cell numbers were calculated using CountBright absolute counting beads (Life Technologies, Grand Island, NY; C36950) according to manufacturer’s instructions.
Immunofluorescence staining of GATA-3
IL44get mice were infected with N. brasiliensis, and lungs, mediastinal lymph nodes, and spleens were harvested 8 days after infection. Single-cell suspensions were prepared and GFP+ CD4+ T cells, ILC2, and basophils were sorted from the lung. GFP+ CD4+ T cells were sorted from the mediastinal lymph nodes, and GFP+ NKT cells were sorted from the spleens of infected mice. Staining for GATA-3 was performed as previously described.57 All following steps were carried out at room temperature. Freshly sorted cells were seeded onto poly-L-lysine-coated slides for 30 min; fixed in 2% formaldehyde in PBS for 10 min; and followed by permeabilization in 0.2% Triton X-100 in PBS for 30 min. The fixed and permeabilized cells were then blocked in 10% goat serum in PBS for 20 min. The cells were then stained with mouse monoclonal anti-human GATA-3 IgG1 (HG3-31, Santa Cruz Biotechnology, Dallas, TX) in blocking buffer, for 1 h in the dark. Cells were subsequently stained with goat anti-mouse Alexa Fluor 555 (Life Technologies; A21127) for 45 min in the dark, and counterstained with 4′,6-diamidino-2-phenylindole, dihydrochloride for 5 min. Images were obtained using the Zeiss AxioImager fluorescence microscope, and analyzed using Adobe Photoshop (Adobe Systems Incorporated, San Jose, CA).
In vitro NKT culture
NKT cells were seeded into U-bottom 96-well plates and cultured with 10 ng ml−1 IL-2 and 10 μM MG132 (Sigma-Aldrich, St. Louis, MO; M7449) for 12 h at 37 °C.
Statistical analysis
Statistical significance was determined by two-tailed unpaired t-tests where *P<0.05, **P<0.005, and ***P<0.0001.
References
Pulendran, B. & Artis, D. New paradigms in type 2 immunity. Science 337, 431–435 (2012).
Locksley, R.M. Asthma and allergic inflammation. Cell 140, 777–783 (2010).
Fallon, P.G. et al. IL-4 induces characteristic Th2 responses even in the combined absence of IL-5, IL-9, and IL-13. Immunity 17, 7–17 (2002).
Liang, H.E., Reinhardt, R.L., Bando, J.K., Sullivan, B.M., Ho, I.C. & Locksley, R.M. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 13, 58–66 (2012).
Grunig, G. et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science 282, 2261–2263 (1998).
Wills-Karp, M. et al. Interleukin-13: central mediator of allergic asthma. Science 282, 2258–2261 (1998).
Bancroft, A.J., McKenzie, A.N. & Grencis, R.K. A critical role for IL-13 in resistance to intestinal nematode infection. J. Immunol. 160, 3453–3461 (1998).
Paget, C. & Trottein, F. Role of type 1 natural killer T cells in pulmonary immunity. Mucosal Immunol. 6, 1054–1067 (2013).
Middendorp, S. & Nieuwenhuis, E.E. NKT cells in mucosal immunity. Mucosal Immunol. 2, 393–402 (2009).
Lisbonne, M. et al. Cutting edge: invariant V alpha 14 NKT cells are required for allergen-induced airway inflammation and hyperreactivity in an experimental asthma model. J. Immunol. 171, 1637–1641 (2003).
Akbari, O. et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat. Med. 9, 582–588 (2003).
Matsuda, H. et al. Plasticity of invariant NKT cell regulation of allergic airway disease is dependent on IFN-gamma production. J. Immunol. 185, 253–262 (2010).
Cui, J. et al. Inhibition of T helper cell type 2 cell differentiation and immunoglobulin E response by ligand-activated Valpha14 natural killer T cells. J. Exp. Med. 190, 783–792 (1999).
Chandra, S. & Kronenberg, M. Activation and function of iNKT and MAIT cells. Adv. Immunol. 127, 145–201 (2015).
Iwamura, C. & Nakayama, T. Role of NKT cells in allergic asthma. Curr. Opin. Immunol. 22, 807–813 (2010).
Thomas, S.Y., Chyung, Y.H. & Luster, A.D. Natural killer T cells are not the predominant T cell in asthma and likely modulate, not cause, asthma. J. Allergy Clin. Immunol. 125, 980–984 (2010).
Akbari, O. et al. CD4+ invariant T-cell-receptor+ natural killer T cells in bronchial asthma. N. Engl. J. Med. 354, 1117–1129 (2006).
Stetson, D.B. et al. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J. Exp. Med. 198, 1069–1076 (2003).
Matsuda, J.L. et al. Mouse V alpha 14i natural killer T cells are resistant to cytokine polarization in vivo. Proc. Natl. Acad. Sci. USA 100, 8395–8400 (2003).
Michel, M.L. et al. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J. Exp. Med. 204, 995–1001 (2007).
Coquet, J.M. et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1-NKT cell population. Proc. Natl. Acad. Sci. USA 105, 11287–11292 (2008).
Matsuda, J.L., Mallevaey, T., Scott-Browne, J. & Gapin, L. CD1d-restricted iNKT cells, the 'Swiss-Army knife' of the immune system. Curr. Opin. Immunol. 20, 358–368 (2008).
Watarai, H. et al. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 10, e1001255 (2012).
Lee, Y.J., Holzapfel, K.L., Zhu, J., Jameson, S.C. & Hogquist, K.A. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14, 1146–1154 (2013).
Carr, T., Krishnamoorthy, V., Yu, S., Xue, H.H., Kee, B.L. & Verykokakis, M. The transcription factor lymphoid enhancer factor 1 controls invariant natural killer T cell expansion and Th2-type effector differentiation. J. Exp. Med. 212, 793–807 (2015).
Pellicci, D.G., Hammond, K.J., Uldrich, A.P., Baxter, A.G., Smyth, M.J. & Godfrey, D.I. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J. Exp. Med. 195, 835–844 (2002).
Benlagha, K., Kyin, T., Beavis, A., Teyton, L. & Bendelac, A. A thymic precursor to the NK T cell lineage. Science 296, 553–555 (2002).
Benlagha, K., Wei, D.G., Veiga, J., Teyton, L. & Bendelac, A. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202, 485–492 (2005).
Dickgreber, N. et al. Immature murine NKT cells pass through a stage of developmentally programmed innate IL-4 secretion. J. Leukoc. Biol. 92, 999–1009 (2012).
Price, A.E. et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. USA 107, 11489–11494 (2010).
Mohrs, M., Shinkai, K., Mohrs, K. & Locksley, R.M. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity 15, 303–311 (2001).
Mohrs, K., Wakil, A.E., Killeen, N., Locksley, R.M. & Mohrs, M. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity 23, 419–429 (2005).
Jin, C. et al. Particulate allergens potentiate allergic asthma in mice through sustained IgE-mediated mast cell activation. J. Clin. Invest. 121, 941–955 (2011).
Ouyang, W. et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity 12, 27–37 (2000).
Oliver, P.M. et al. Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 25, 929–940 (2006).
Yamashita, M. et al. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J. Biol. Chem. 280, 29409–29419 (2005).
Fang, D. et al. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3, 281–287 (2002).
Jin, H.S., Park, Y., Elly, C. & Liu, Y.C. Itch expression by Treg cells controls Th2 inflammatory responses. J. Clin. Invest. 123, 4923–4934 (2013).
Serafini, N. et al. Gata3 drives development of RORgammat+ group 3 innate lymphoid cells. J. Exp. Med. 211, 199–208 (2014).
Gessner, A., Mohrs, K. & Mohrs, M. Mast cells, basophils, and eosinophils acquire constitutive IL-4 and IL-13 transcripts during lineage differentiation that are sufficient for rapid cytokine production. J. Immunol. 174, 1063–1072 (2005).
Loots, G.G. et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288, 136–140 (2000).
Kelly, B.L. & Locksley, R.M. Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J. Immunol. 165, 2982–2986 (2000).
Dolganov, G. et al. Coexpression of the interleukin-13 and interleukin-4 genes correlates with their physical linkage in the cytokine gene cluster on human chromosome 5q23-31. Blood 87, 3316–3326 (1996).
Kishikawa, H., Sun, J., Choi, A., Miaw, S.C. & Ho, I.C. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J. Immunol. 167, 4414–4420 (2001).
Zhang, D.H., Yang, L. & Ray, A. Differential responsiveness of the IL-5 and IL-4 genes to transcription factor GATA-3. J. Immunol. 161, 3817–3821 (1998).
Zhu, J. et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat. Immunol. 5, 1157–1165 (2004).
Mjosberg, J. et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity 37, 649–659 (2012).
Hoyler, T. et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 37, 634–648 (2012).
Wang, Z.Y., Kusam, S., Munugalavadla, V., Kapur, R., Brutkiewicz, R.R. & Dent, A.L. Regulation of Th2 cytokine expression in NKT cells: unconventional use of Stat6, GATA-3, and NFAT2. J. Immunol. 176, 880–888 (2006).
Zheng, W. & Flavell, R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997).
Yamashita, M. et al. Essential role of GATA3 for the maintenance of type 2 helper T (Th2) cytokine production and chromatin remodeling at the Th2 cytokine gene loci. J. Biol. Chem. 279, 26983–26990 (2004).
Klein Wolterink, R.G. et al. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc. Natl. Acad. Sci. USA 110, 10240–10245 (2013).
Barlow, J.L. et al. IL-33 is more potent than IL-25 in provoking IL-13-producing nuocytes (type 2 innate lymphoid cells) and airway contraction. J. Allergy Clin. Immunol. 132, 933–941 (2013).
Saenz, S.A. et al. IL-25 simultaneously elicits distinct populations of innate lymphoid cells and multipotent progenitor type 2 (MPPtype2) cells. J. Exp. Med. 210, 1823–1837 (2013).
Corren, J. et al. Lebrikizumab treatment in adults with asthma. N. Engl. J. Med. 365, 1088–1098 (2011).
Voehringer, D., Reese, T.A., Huang, X., Shinkai, K. & Locksley, R.M. Type 2 immunity is controlled by IL-4/IL-13 expression in hematopoietic non-eosinophil cells of the innate immune system. J. Exp. Med. 203, 1435–1446 (2006).
Frelin, C. et al. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat. Immunol. 14, 1037–1044 (2013).
Acknowledgements
We thank Xiaoping Zhong and Tiffany Carr (Duke University) for expert review and comments; Lynn Martinek and Ann Miller for technical expertise and support. We also thank the NIH tetramer core for graciously providing CD1d-tetramer reagents used in these studies. This work was supported by the National Institutes of Health (AI119004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Additional information
Author contributions
T.F.O. and R.L.R. conceived the work. T.F.O., R.L.R., K.B., M.D., W.X.G.A, and S.A. designed and/or performed experiments. T.F.O, K.B., and R.L.R wrote the manuscript.
SUPPLEMENTARY MATERIAL is linked to the online version of the paper
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit https://doi.org/creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
O'Brien, T., Bao, K., Dell'Aringa, M. et al. Cytokine expression by invariant natural killer T cells is tightly regulated throughout development and settings of type-2 inflammation. Mucosal Immunol 9, 597–609 (2016). https://doi.org/10.1038/mi.2015.78
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mi.2015.78
This article is cited by
-
Thymic development of unconventional T cells: how NKT cells, MAIT cells and γδ T cells emerge
Nature Reviews Immunology (2020)
-
Thymic iNKT single cell analyses unmask the common developmental program of mouse innate T cells
Nature Communications (2020)
-
Egg yolk augments type 2 immunity by activating innate cells
European Journal of Nutrition (2020)
-
Distinct gene expression patterns correlate with developmental and functional traits of iNKT subsets
Nature Communications (2016)