Abstract
The nucleoside analog azacitidine (AZA) is used in the treatment of myelodysplastic syndromes (MDS), but 30–40% of patients fail to respond or relapse after treatment. Hence, to identify new molecular alterations that allow for identification of patients unlikely to respond to AZA could impact the utility of this therapy. We determined the expression levels of genes involved in AZA metabolism: UCK1, UCK2, DCK, hENT1, RRM1 and RRM2 using quantitative PCR in samples from 57 patients with MDS who received AZA. Lower expression of UCK1 was seen in patients without a response to AZA (median 0.2 vs 0.49 for patients with response to AZA, P=0.07). This difference in UCK1 expression was not influenced by aberrant methylation of the UCK1 promoter. In addition, the seven polymorphic loci found in the coding sequence were not associated with UCK1 gene expression nor AZA response. Silencing of UCK1 by siRNA leads to blunted response to AZA in vitro. The univariate analysis revealed that patients expressing lower than median levels of UCK1 had a shorter overall survival (P=0.049). Our results suggest that expression level of UCK1 is correlated with clinical outcome and may influence the clinical response to AZA treatment in patients with MDS.
Similar content being viewed by others
Introduction
The hypomethylating agent azacitidine (AZA) is the standard therapy for the treatment of patients with higher-risk myelodysplastic syndromes (MDS) (International Prognostic Scoring System (IPSS) intermediate-2 or high) and patients with acute myeloid leukemia (AML) with 20–30% bone marrow (BM) blasts in Europe, while in the United States it is approved for all French–American–British (FAB) MDS subtypes. Although AZA is extensively used, 30–40% of patients fail to respond or relapse after treatment.1, 2
There have been efforts to characterize biological and clinical parameters predictive of response or resistance to AZA, and although some have been identified, they have not yet been validated to support clinical decisions.3, 4, 5, 6 The concept that level of DNA methylation may correlate with response to treatment has been considered for some time, but there is not sufficient evidence to fully confirm this hypothesis as few data are available. For example, an initial study focused on CDKN2B methylation, but this did not correlate with response.3 Similarly, phosphoinositide/phospholipase C beta 1 was shown to be hypermethylated in patients with MDS and reduction of methylation was correlated with response to AZA.4, 7 Moreover, Shen et al.8 showed that reduced global methylation over time correlates with survival and response to therapy in patients with MDS.
Several gene mutations that correlate with prognosis in patients with MDS have been evaluated for their power to predict response to AZA. For example, TET2 mutations correlated with response to AZA but without impacting response duration or survival.9 Mutations in other genes involved in epigenetic regulation, including DNMT3A, ASXL1, TET2 and IDH1/IDH2, may impact response to AZA.10 It has also been shown that resistance to AZA significantly correlated with the percentage of MDS or AML cells expressing the BCL-2-like protein 10.5 Finally, clinical parameters have been identified that independently predicted lower response rates (previous low-dose cytosine arabinoside (Ara-C), BM blasts>15% and abnormal karyotype) and reduced overall survival (OS) (performance status ⩾2, intermediate- and poor-risk cytogenetics, presence of circulating blasts and red blood cell transfusion dependency with AZA) in patients with higher-risk MDS.11
AZA is a nucleoside analog (NA) that mimics physiological cytidine in terms of uptake into the cell and activation to exert its pharmacological action.12, 13 The concentrative nucleoside transporter and equilibrative nucleoside transporter proteins mediate the transport of NAs such as AZA across membranes.14, 15 Inside cells, AZA is activated by the enzyme uridine-cytidine kinase (UCK).13, 16 The majority (80–90%) of activated AZA is incorporated into RNA resulting in mRNA and protein metabolism disruption.17, 18, 19 A smaller fraction (10–20%) of AZA is converted to decitabine (DAC) by ribonucleotide reductase (RR) and incorporated into newly synthesized DNA, depleting DNA methyltransferase 1 and leading to DNA hypomethylation.15, 17
Many of the mechanisms of primary resistance to NAs are related to alterations in components that impact their metabolism.20, 21, 22, 23, 24 Mutations in and/or expression levels of nucleoside-activating or inactivating enzymes have been correlated with tumor response or patient outcome with Ara-C.22, 23, 24, 25, 26 A recent work indicated that increased cytidine deaminase (CDA; enzyme inactivating NA) expression/activity in plasma of male MDS patients contributes to decreased drug plasma half-life and could account for worse outcomes with NA therapy.27 We hypothesized that alterations in AZA cellular uptake and metabolism could be a mechanism for relapse or refractoriness to treatment.
We studied the expression of genes encoding nucleoside transporter proteins and enzymes involved in the metabolism of AZA: UCK1 and UCK2, deoxycytidine kinase (DCK), human equilibrative nucleoside transporter 1 (hENT1) and the two subunits of the RR gene (RRM1 and RRM2) in samples from primary MDS patients and correlated these data with response to AZA and clinical outcomes.
Patients and methods
Patients
The study population included 57 patients diagnosed with MDS or AML with 20–30% blasts between June 2006 and August 2012. MDS and AML diagnosis was made in accordance with the World Health Organization (WHO) criteria.28 Three chronic myelomonocytic leukemia cases were diagnosed according to FAB morphological and cytogenetic criteria29 and were included in the study. BM mononuclear cells were collected at diagnosis or immediately before AZA treatment separated on Ficoll Hypaque density gradient (Lympholyte-H; Cedarlane Labs, Hornby, ON, Canada). Informed consent was provided in accordance with the Declaration of Helsinki Principles and local Ethics Committee’s approval. BM mononuclear cells from 10 healthy donors obtained after informed consent were also separated as negative controls.
AZA was given subcutaneously following the schedule approved by the US Food and Drug Administration and European Medicines Agency of 75 mg/m2 per day for 7 days every 28 days. Response was analyzed on an intention-to-treat basis. Patients received at least two cycles of AZA and underwent full BM and hematological evaluation to determine response according to the International Working Group criteria for MDS30 and AML.31 Patients were considered responders when in complete remission (CR), marrow CR (mCR), partial remission (PR) or hematological improvement (HI) and non-responders when in stable disease (SD) or progressive disease (PD). Characteristics of all patients are described in Table 1. Though the IPSS score validation was stated for untreated patients with primary MDS, patients who had received growth factors or short courses (⩽3 months) of low-dose chemotherapy were included in the analysis.32 Herein, IPSS score was recalculated before the onset of AZA therapy.
Expression analysis
Quantitative PCR (qPCR) was carried out with the ABI GeneAmp 5700 instrument and software (Applied Biosystems, Foster City, CA, USA). Specific oligonucleotides and TaqMan probes were available from Applied Biosystems (catalog no: UCK1: Hs01075618_m1; UCK2: Hs:00367072_m1, DCK: Hs00176127_m1, hENT1: Hs01085706_m, RRM1: Hs01040698_m1, RRM2: Hs01072069_g1). All qPCR experiments were normalized to the housekeeping gene glyceraldehyde-2-phosphate dehydrogenase (GAPDH). Each reaction was performed in duplicate.
The mean value of the replicates for each sample was calculated and expressed as threshold cycle (CT) values. The amount of gene expression was measured as the difference (ΔCT) between the CT value of the sample for the target gene and the CT value of the sample for the GAPDH endogenous control gene (ΔCT=CT(Target)−CT(GAPDH)). The results were then expressed as 2−(ΔCt).
Methylation-specific PCR
One microgram of genomic DNA was converted with bisulfite before amplification. After conversion, methylation of UCK1 was assessed by methylation-specific PCR using specific primers (Supplementary Table S1). DNA from mononuclear cells from healthy donors was used as a negative control and methylated control DNA (CpGenome Universal Methylated DNA; Intergen, Purchase, NY, USA) was used as positive control. The PCR conditions were: hot start at 95 °C for 10 min followed by 40 cycles at 95 °C for 30 s, 56 °C for 1 min, and 72 °C for 45 s, followed by a final extension at 72 °C for 10 min.
Sequencing analysis
The complete coding sequence of the UCK1 gene was sequenced by the Sanger method. Five PCR amplicons were designed in order to encompass all the exonic regions within the UCK1 gene, including at least 50 base pairs at the intron/exon boundaries. Primers are listed in Supplementary Table S1. The reaction products were loaded onto an ABI prism 3100 Genetic analyzer (Applied Biosystems). Primary sequencing data were analyzed using the Geneious software (Biomatters Ltd, Auckland, New Zealand) and compared with the human UCK1 gene sequence and its complete coding sequence (Gene ID: 83549, CCDS6944).
When enough DNA was available, standard PCR and sequencing of the entire coding sequence (exons 3–11) of the TET2 gene were performed in BM genomic DNA, as previously described.33
siRNA transfection
For UCK1 silencing, two siRNAs were used: SR313172C (siUCK1-1) (AUAAUGAUUUGAUGCACAGGACUCT) and SR3131172B (siUCK1-2) (GGUUUAAAGAUCCCUCUAGGUCACT) (OriGene Technologies, Inc., Rockville, MD, USA). A designated non-targeting siRNA, scramble SR30005 (SCR), provided a control for non-specific siRNA transfection effects on gene expression. K562 cells were cultured at a density of 1 × 106 cells/ml in 6 ml of RPMI 1640 medium. The TransIT-TKO transfection reagent was diluted in medium free of antibiotics and the siRNAs in SR30005-Rnase-free siRNA duplex re-suspension buffer, and then the two components were mixed. After 30 min of incubation, K562 cells were cultured with the siRNAs, the TKO transfectant reagent and the SCR. After 48 h of transfection, cells were treated for further 24 h with AZA at the optimal concentration of 1 μM as previously established in our laboratory.34 After treatment, cells were washed, and RNA and proteins were extracted. Each experiment was performed in duplicate. Effective UCK1 silencing was evaluated by qPCR with the method described above.
Western blotting
The effective presence and downregulation of the UCK1 protein was evaluated by western blotting. The protein extracts of the K562 cultures were studied using the following antibodies: anti-UCK1 mouse monoclonal antibody (1:1000, OriGene Technologies) and the antibody against α-tubulin (1:1000; Sigma-Aldrich, St Louis, MO, USA) used to confirm equal loading of extracts.
Annexin-V-binding assay
To quantify apoptosis, cells were washed twice with phosphate-buffered saline, and the cell pellet was resuspended in 100 ml of Annexin-V-FLUOS (HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffered saline solution with 2.5 mM CaCl2). Phycoerythrin-labeled Annexin-V (Roche Diagnostics GmbH, Mannheim, Germany) and propidium iodide (PI) were added, and cells were incubated for 15 min in the dark at room temperature. After the addition of 400 ml of binding buffer and agitation, flow cytometer analysis was performed using a FACSscan (Becton Dickinson, Franklin Lakes, NJ, USA). Cells positively stained with Annexin-V and PI-negative were considered apoptotic.
Statistical analysis
All descriptive statistics and tests (Mann–Whitney nonparametric U-test, Kruskal–Wallis test, Chi-square test and Fisher’s exact test) were performed using the statistical package SPSS, version 17.0 (SPSS Inc., Chicago, IL, USA). Regarding the polymorphic loci of UCK1, unconditional logistic regression was used to assess genotype distributions between patients with or without response to AZA, considering the homozygotes for the more frequent allele as the reference class. Due to the small numbers of samples available, heterozygous and homozygous subjects for the variant alleles were grouped together. A P-value ⩽0.05 was considered statistically significant.
OS was defined as the time from initiation of AZA treatment to death from any cause. Patients who remained alive were censored at the time of last follow-up. Survival from time of MDS diagnosis was also evaluated in patients with or without response to AZA. Time-to-event curves were estimated by the Kaplan–Meier method and compared by stratified log rank tests. Stratified Cox proportional hazards regression models were used to estimate hazard ratios (HRs) and associated 95% confidence intervals. The primary analysis of OS for patients with or without response to AZA used the stratified Cox proportional hazards model without any covariate adjustments to estimate the HR. All P-values reported are two sided.
Results
Patient characteristics
This study included samples from 57 patients recruited from three different centres in Italy (36/21 M/F with a median age at diagnosis of 70 years (range, 37–86 years). At baseline, median hemoglobin level, absolute neutrophil count and platelet count were 8.9 g/dl (range, 6–14.1), 2 × 109 cells/l (range, 1 × 109–15 × 109) and 48 × 109/l (range, 3 × 109–267 × 109), respectively. Median BM blast count was 13.2%. Cytogenetic risk was good in 35 (64%), intermediate in 6 (11%) and poor in 14 (25%) patients. IPSS was intermediate-1 in 7 (13%), intermediate-2 in 33 (62%) and high in 13 (24%) patients. WHO diagnosis included 4 (7%) cases of refractory anemia (RA), RA with ringed sideroblasts or refractory cytopenia with multilineage dysplasia, 12 (22%) cases of RA with excess of blasts-1, 25 (46%) cases of RA with excess of blasts-2 and 10 (18%) cases of RA with excess of blasts in transformation/AML. Three patients (7%) were diagnosed with chronic myelomonocytic leukemia according to the FAB classification.
The median number of cycles of AZA received was 8 (range, 2–47). Of the 57 patients, 23 (40%) achieved a response to AZA (CR, PR, HI or mCR) and 34 (60%) did not (SD or PD) (Table 2). Overall, median OS from initiation of AZA therapy was 27.2 months; patients who achieved CR/mCR, PR or HI had a median OS of 54.1, 38 and 23.1 months, respectively, confirming that OS was longer in patients with CR/PR/HI, compared with patients with no response, as previously published.2 In fact, the median OS from diagnosis in patients with or without a response to AZA was 72.3 and 35.1 months, respectively. When the scoring system designed by Itzykson et al.11 for predicting OS after AZA was applied to our MDS patients, 18% of them were included in the low-, 79% in the intermediate- and 3% in the high-risk category.
Analysis of TET2 mutations
Eleven TET2 mutations were found in eight of 35 patients, including three frameshift mutations (all inducing a premature STOP codon), three nonsense and five missense mutations, all five in the first conserved domain and one of them was found in homozygosis. Three patients showed two different mutations in the gene. Among patients with TET2 mutations, three patients showed a response to AZA (CR, n=1; PR, n=1; and HI, n=1) while five patients did not (SD, n=3; PD, n=2) (P=0.7).
Expression levels of enzymes involved in the metabolism of AZA in MDS patients
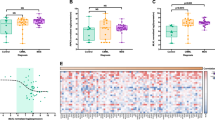
Gene expression of the studied enzymes was heterogeneous among MDS cases, and the level of mRNA expression was different for the various genes (UCK1, median 0.25, Q1: 0.15–Q3: 0.55; UCK2, median 0.45, Q1: 0.25–Q3: 0.76; DCK, median 5.2, Q1: 3.15–Q3: 10.9; hENT1, median 5.9, Q1: 2.7–Q3: 12.6; RRM1 median 1.3, Q1: 0.6–Q3: 2.6; and RRM2 median 9.2, Q1: 3.7–Q3: 20.1; Figure 1a). We investigated the association between the expression of each gene and the response to AZA treatment as well as other clinical characteristics. Generally, the expression level of each gene was not significantly different in patients with or without response to AZA. However, lower expression of the UCK1 gene was shown in patients without response to AZA (median 0.23 (Q1: 0.15–Q3: 0.40) vs median 0.49 (Q1: 1.7–Q3: 9.3); P=0.07) (Figure 1b). We also evaluated the expression of UCK1 in the three different groups of the AZA scoring system,11 observing no differences among them. Mean UCK1 expression level in the low-risk group was 0.28±0.13, in the intermediate 0.45±0.38 and in the high 0.15 (P=0.22). No correlation was found with age, sex, hemoglobin, absolute neutrophil count, platelet count, WHO classification, cytogenetic risk group, TET2 mutations, IPSS risk groups or BM blast percentage for expression of UCK1, UCK2, DCK, hENT1, RMM1 or RMM2 (data not shown).
Expression of genes implicated in the metabolism of AZA. (a) Expression of UCK1, UCK2, DCK, hENT1, RRMI and RRM2 (relative to GAPDH) in patients with MDS (n=57) before AZA treatment. (b) Expression of UCK1 (relative to GAPDH) in patients without response to AZA (SD or PD) vs patients with response to AZA (CR, PR, HI and mCR). In both plots, the thick middle line represents the median expression of the different genes, and the top and bottom edges of the box represent the 75th and 25th percentiles, respectively. The outliers are also represented by open and filled circles and asterisks.
Molecular alterations of UCK1 in MDS patients
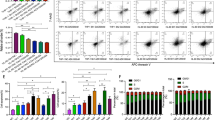
In order to investigate the molecular events leading to differential expression of UCK1, we measured the methylation status of the CpG island at the UCK1 promoter. The CpG island was unmethylated in all the samples analyzed and in healthy controls (Figure 2a). To assess whether the difference in UCK1 expression could be due to the presence of genetic variants, we sequenced the UCK1 coding sequence in a subset of 42 patients for which genomic DNA was available. By this method, seven polymorphic loci were identified. The position of the loci was compared with resequencing data reported in the 1000 Genomes project (http://www.1000genomes.org/). All seven loci corresponded to known single-nucleotide polymorphism, three of which fell into intronic sequences (rs3904960 in Intron 1; rs2296956 and rs189473964 in Intron 3), two in regulatory sequences (rs7041225 in the 5′ untranslated region; rs150900763 in the 3′ untranslated region) and two in the coding sequences of exon 2 (rs7867616, Glu29Gln) and exon 6 (rs2296957, Ala180Ser) (Figure 2b). For the SNPs rs189473964 and rs150900763, the variant allele was present in only one subject, and these two SNPs were therefore excluded from any further analysis. None of the remaining five variants were shown to affect UCK1 expression levels (Figure 2c). Association of genotypes with achievement of response to AZA was also assessed, but none were found.
Molecular alterations of UCK1 in MDS patients. (a) Representative examples of methylation-specific PCR analysis of UCK1 in seven MDS cases without response to AZA and six cases of MDS patients with response to AZA. C(+), DNA universally methylated; C(−), healthy individual; blank, control without added DNA; U, unmethylated alleles; and M, methylated. (b) UCK1 gene locus and the sequenced regions. Five PCR were designed to sequence the complete coding DNA sequence (Exon 1, Exon 2, Exon 3-5, Exon 6 and Exon 7) for all the isoforms of the UCK1 protein. A total of seven common genetic variants, indicated by their reference number, were found to be polymorphic in at least one subject. (c) UCK1 expression levels in different genotype groups. Gene expression values were compared with the genotypes of the genetic variants identified. In the plots, the thick middle line represents the median expression of the different genes, and the top and bottom edges of the box represent the 75th and 25th percentiles, respectively. The outliers are represented by asterisks.
Silencing of UCK1 gene expression
To understand the effects of UCK1 on the sensitivity to AZA, we carried out a knockdown experiment using two synthetic siRNAs against the UCK1gene. K562 cells cultured in the presence of two siRNAs for 48 h plus 24 h of exposure to AZA showed a significant downregulation of the UCK1 mRNA compared with control cultures with only one of the two siRNA tested (siUCK1-1 vs control, P=0.046; siUCK1-1+AZA vs control+AZA, P=0.021; Figure 3a). UCK1 protein expression was robustly diminished in K562 cells exposed to siUCK1-1 (Figure 3b). When UCK1 was downregulated, a reduction of AZA-induced apoptosis was observed (siUCK1-1: Annexin V-positive cells 11%±0.34% vs non-silenced control cultures: Annexin V-positive cells 31%±0.65%, P=0.0015; Figure 3c). siUCK1-2 was not able to silence the gene, and in that case, there was no variation in the sensitivity to AZA with respect to control or mock transfected cells (Supplementary Figure S1).
Silencing of the UCK1 gene. Effect of siUCK1-1 on the expression of the UCK1 gene (relative to GAPDH) was evaluated using quantitative PCR (a) and western blotting analysis (b). Downregulation of the UCK1 mRNA by siUCK1-1 was observed in non-transfected compared with mock transfected control cells, both not treated (*P=0.046) and treated with AZA (**P=0.021) (a). Mean of two different experiments is shown. UCK1 protein expression was equally diminished in K562 cells exposed to siUCK1-1 (b). (c) Apoptosis (Annexin V-FITC (fluorescein isothiocyanate) and PI staining) was measured in K562 cells cultured with SRC, TKO and siUCK1-1, without AZA and with 24-h AZA. AZA induced an increase in Annexin V-positive cells, but apoptosis was blunted in K562 cells when siUCK1-1 was present and UCK1, as well as UCK1 protein, were downexpressed (*P=0.0015, siUCK1-1 transfected vs control culture).
Prognostic impact of clinical characteristics and correlation with UCK1 expression
To assess the variables predictive of OS from onset of AZA treatment, we analyzed the baseline characteristics of sex, age, BM blast percentage, IPSS risk score, cytogenetics (within IPSS subgroups), score according to ‘AZA scoring system’,11 TET2 mutations and the different genotypes found in the UCK1 gene. Univariate analysis showed that OS was significantly influenced only by IPSS risk score (P=0.02) and achievement of response to AZA (P=0.001). Median OS by the ‘AZA scoring system’ was not reached in the low-risk group vs 44.7 months in the intermediate-risk group vs 14.9 months in the high-risk group (P=0.18). We also evaluated in the univariate analysis the prognostic impact of the expression of UCK1, UCK2, DCK, hENT1, RRM1 and RRM2. The median levels of gene expression were used to divide cases with low (below median) or high (above median) expression for each gene. We demonstrated that patients with lower expression of UCK1 had shorter OS than patients with higher expression of UCK1 (median OS values: 19 months vs 49 months; P=0.041; Figure 4). Levels of UCK2, DCK, hENT1, RRM1 and RRM2 expression did not significantly impact OS. Multivariate analysis showed that the independent factors influencing OS were achievement of response to AZA (HR, 0.23; P=0.0016) and higher expression of UCK1 (HR, 0.38; P=0.049).
Discussion
This study suggests that the expression of UCK1, involved in the activation of AZA, may influence the clinical response to AZA treatment in patients with MDS. The biological regulation of UCK1 expression was not due to methylation status of the UCK1 promoter nor to the presence of genetic polymorphisms in the UCK1 sequence. Prolonged OS in this series of AZA-treated patients with MDS was associated with higher expression of UCK1.
Hypomethylating agents AZA and DA have been shown to induce overall responses in approximately 40–60% of patients with MDS.1, 2, 35, 36 However, only AZA has been shown to significantly prolong OS in patients with IPSS higher-risk MDS. In our cohort, we confirmed that response to AZA is correlated with prolonged survival. There have been efforts to elucidate factors predictive of response to AZA, in hopes of selecting candidates most likely to respond to treatment. We applied the ‘AZA scoring system’11 to our cohort of MDS patients, and although we could stratify them for OS, only a trend to longer survival was observed for low scoring ones (9/57), the vast majority belonging to intermediate scoring group. Both AZA and DAC inhibit DNA methyltransferase activity and methylation of new DNA strands. MDS is clinically the most responsive disease to these agents, potentially due to its frequent aberrant methylation.8, 37 The correlation between the presence of mutations in several epigenetic interfering genes (for example, TET2, EZH2, ASXL1 and DNMT3) and response to AZA has been shown but must be further confirmed to determine clinical applicability.9, 10, 38 Only eight of the 35 MDS patients analyzed in our study had mutated TET2. As expected, there was no correlation with mutational status and OS, but we could not observe higher rates of response to AZA in mutated cases, most probably because of numerical limitations.
Although AZA and DAC have been shown to induce DNA hypomethylation in vitro and in vivo,19 despite some reports,8 research has failed to elucidate the correlation between baseline DNA hypermethylation and response to AZA or DAC. Therefore, the identification of biological markers that permit the selection of patients with MDS unlikely to respond to AZA therapy is of great importance.
The cytosine NAs AZA and DAC are transported inside the cell via specific proteins and then converted into active nucleotides to induce DNA hypomethylation and cytotoxicity.39, 40 An important mechanism of resistance to NAs may be low levels of active intracellular agent resulting from insufficient cellular uptake, reduced levels of activating enzymes and increased degradation of the agents. Therefore, we speculated that similar mechanisms could account for resistance to AZA. The first ATP-dependent phosphorylation, catalyzed by the enzyme UCK for AZA and DCK for DAC,39, 40 is a limiting step for AZA and DAC activation. In particular, DCK deficiency has been reported as a major mechanism of resistance to the NAs DAC, Ara-C and gemcitabine.21, 22, 41, 42 Recently, it has been reported that higher plasma levels of CDA may account for worse outcome after NA treatment in male MDS patients.27 In that study, CDA production by the liver and its presence in plasma were evaluated, analyzing its impact on DAC pharmacokinetics. We did not observe any difference in AZA response rates between male and female patients. On the other hand, there is still limited information regarding the relationship between alterations in the components of cellular AZA metabolism and the resistance seen in 30–40% of AZA-treated patients with MDS.1, 2 AZA incorporation into RNA may enhance its clinical activity in MDS, as survival data suggest superiority to DAC, which only undergoes DNA incorporation.13
We compared the expression of UCK1, UCK2, DCK, hENT1, RRM1 and RRM2 in MDS patients with or without clinical response to AZA. Lower expression levels of UCK1 were shown in patients resistant to AZA. In contrast to UCK1, expression levels of UCK2 were not significantly different in responsive vs non-responsive patients. One may presume that both proteins phosphorylate AZA to its nucleoside monophosphate, but our results suggest that these two enzymes could act independently and do not seem to have the same relevance in AZA metabolism. In MDS patients with low expression of UCK1, we did not observe a compensatory higher expression of UCK2. Consistently with our finding, it has been demonstrated that the phosphorylation efficiency of the enzymes UCK1 and UCK2 on the same substrate can be different.43 Levels of DCK, hENT1, RRM1 and RRM2 expression were not associated with achievement of response to AZA. Additionally, we also found that OS from onset of AZA treatment was significantly shorter in patients with lower levels of UCK1 expression. The prognostic weight of UCK1 may reflect a mechanism of action specific to AZA, which modulates protein synthesis via RNA incorporation. RRM1 and RRM2 did not show lower expression levels in patients without response to AZA in this study. This could suggest that the step catalyzed by RRM1 and RRM2, which converts AZA to DAC and leads to DNA incorporation,18 is not discriminative for the clinical activity of this agent.
Aberrant hypermethylation of UCK1 was not present in any patient examined, including healthy controls. In addition, the various polymorphisms found in the UCK1 sequence were not related to its expression level nor with response to AZA treatment, alternate to what has been demonstrated for DCK.44 We did not explore other possible mechanisms of gene expression regulation, such as miRNA or splicing variants.45 However, UCK1 has several regulating miRNAs, (http://www.targetscan.org/) and future studies are warranted, because miRNAs could be helpful tools to modulate sensitivity to AZA. In fact, in order to explore this hypothesis and support our findings, we elaborated a functional assay, silencing UCK1 by siRNA. In the in vitro model, the reduced expression of UCK1 in the leukemic cell line K562 decreased its otherwise significant AZA-induced apoptosis. Although other variables may impact response in the clinical setting, these results highlight the importance of UCK1 for AZA activity.
Although we demonstrated that level of UCK1 expression is correlated with outcome to AZA therapy, low expression of DCK, which phosphorylates DAC, has been shown to impair sensitivity in vitro to DAC.41 Although in hematological neoplasms treated with NAs correlation of DCK levels with clinical outcome remains controversial,46, 47 we may envisage a future where selection of patients with MDS to treat with AZA or DAC is possible on the basis of their levels of UCK1 or DCK, in an attempt to decrease primary resistance. Better definition of the relationship and importance of enzyme expression to clinical response to AZA or DAC in patients with MDS may help to elucidate their specific mechanism of action and further optimize therapy.
References
Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R et al. Randomized, controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the Cancer and Leukemia Group B. J Clin Oncol 2002; 20: 2429–2440.
Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol 2009; 10: 223–232.
Raj K, John A, Ho A, Chronis C, Khan S, Samuel J et al. CDKN2B methylation status and isolated chromosome 7 abnormalities predict responses to treatment with 5-azacytidine. Leukemia 2007; 21: 1937–1944.
Follo MY, Finelli C, Mongiorgi S, Clissa C, Bosi C, Testoni N et al. Reduction of phosphoinositide-phospholipase C beta1 methylation predicts the responsiveness to azacitidine in high-risk MDS. Proc Natl Acad Sci USA 2009; 106: 16811–16816.
Cluzeau T, Robert G, Mounier N, Karsenti JM, Dufies M, Puissant A et al. BCL2L10 is a predictive factor for resistance to azacitidine in MDS and AML patients. Oncotarget 2012; 3: 490–501.
Voso MT, Fabiani E, Piciocchi A, Matteucci C, Brandimarte L, Finelli C et al. Role of BCL2L10 methylation and TET2 mutations in higher risk myelodysplastic syndromes treated with 5-azacytidine. Leukemia 2011; 25: 1910–1913.
Follo MY, Finelli C, Mongiorgi S, Clissa C, Chiarini F, Ramazzotti G et al. Synergistic induction of PI-PLCβ1 signaling by azacitidine and valproic acid in high-risk myelodysplastic syndromes. Leukemia 2011; 25: 271–280.
Shen L, Kantarjian H, Guo Y, Lin E, Shan J, Huang X et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol 2010; 28: 605–613.
Itzykson R, Kosmider O, Cluzeau T, Mansat-De Mas V, Dreyfus F, Beyne-Rauzy O et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia 2011; 25: 1147–1152.
Traina F, Visconte V, Elson P, Tabarroki A, Jankowska AM, Hasrouni E et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia 2014; 28: 78–87.
Itzykson R, Thepot S, Quesnel B, Dreyfus F, Beyne-Rauzy O, Turlure P et al. Prognostic factors for response and overall survival in 282 patients with higher-risk myelodysplastic syndromes treated with azacitidine. Blood 2011; 117: 403–411.
Sorm F, Piskala A, Cihak A, Veselý J . 5-Azacytidine, a new, highly effective cancerostatic. Experientia 1964; 20: 202–203.
Santini V . Azacitidine: activity and efficacy as an epigenetic treatment of myelodysplastic syndromes. Expert Rev Hematol 2009; 2: 121–127.
Rius M, Stresemann C, Keller D, Brom M, Schirrmacher E, Keppler D et al. Human concentrative nucleoside transporter 1-mediated uptake of 5-azacytidine enhances DNA demethylation. Mol Cancer Ther 2009; 8: 225–231.
Pastor-Anglada M, Cano-Soldado P, Molina-Arcas M, Lostao MP, Larráyoz I, Martínez-Picado J et al. Cell entry and export of nucleoside analogues. Virus Res 2005; 107: 151–164.
Lee T, Karon M, Momparler RL . Kinetic studies on phosphorylation of 5-azacytidine with the purified uridine-cytidine kinase from calf thymus. Cancer Res 1974; 34: 2482–2488.
Li LH, Olin EJ, Buskirk HH, Reineke LM . Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer Res 1970; 30: 2760–2769.
Aimiuwu J, Wang H, Chen P, Xie Z, Wang J, Liu S et al. RNA-dependent inhibition of ribonucleotidereductase is a major pathway for 5-azacytidine activity in acute myeloid leukemia. Blood 2012; 119: 5229–5238.
Hollenbach PW, Nguyen AN, Brady H, Williams M, Ning Y, Richard N et al. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS One 2010; 5: e9001.
Galmarini CM, Mackey JR, Dumontet C . Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia 2001; 15: 875–890.
Veuger MJ, Honders MW, Spoelder HE, Willemze R, Barge RM . Inactivation of deoxycytidine kinase and overexpression of P-glycoprotein in AraC and daunorubicin double resistant leukemic cell lines. Leuk Res 2003; 27: 445–453.
Galmarini CM, Thomas X, Graham K, El Jafaari A, Cros E, Jordheim L et al. Deoxycytidine kinase and cN-II nucleotidase expression in blast cells predict survival in acute myeloid leukaemia patients treated with cytarabine. Br J Haematol 2003; 122: 53–60.
Galmarini CM, Graham K, Thomas X, Calvo F, Rousselot P, El Jafaari A et al. Expression of high Km 5'-nucleotidase in leukemic blasts is an independent prognostic factor in adults with acute myeloid leukemia. Blood 2001; 98: 1922–1926.
Hubeek I, Stam RW, Peters GJ, Broekhuizen R, Meijerink JP, van Wering ER et al. The human equilibrative nucleoside transporter 1 mediates in vitro cytarabine sensitivity in childhood acute myeloid leukaemia. Br J Cancer 2005; 93: 1388–1394.
Rathe SK, Largaespada DA . Deoxycytidine kinase is downregulated in Ara-C-resistant acute myeloid leukemia murine cell lines. Leukemia 2010; 24: 1513–1515.
Yin B, Tsai ML, Hasz DE, Rathe SK, Le Beau MM, Largaespada DA . A microarray study of altered gene expression after cytarabine resistance in acute myeloid leukemia. Leukemia 2007; 21: 1093–1097.
Mahfouz RZ, Jankowska A, Ebrahem Q, Gu X, Visconte V, Tabarroki A et al. Increased CDA expression/activity in males contributes to decreased cytidine analog half-life and likely contributes to worse outcomes with 5-azacytidine or decitabine therapy. Clin Cancer Res 2013; 19: 938–948.
Brunning RD, Orazi A, Germing U, Le Beau MM, Porwit A, Baumann I et al. Myelodysplastic syndromes. In: Swederlow SH, Campo E, Lee Harris N, Jaffe ES, Pileri SA, Stein H, et al. (eds) WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. International Agency for Research on Cancer (IARC): Lyon, France, 2008; pp 87–107.
Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol 1982; 51: 189–199.
Cheson BD, Greenberg PL, Bennett JM, Lowenberg B, Wijermans PW, Nimer SD et al. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood 2006; 108: 419–425.
Cheson BD, Bennett JM, Kopecky KJ, Büchner T, Willman CL, Estey EH et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol 2003; 21: 4642–4649.
Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 1997; 89: 2079–2088.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009; 360: 2289–2301.
Buchi F, Spinelli E, Masala E, Gozzini A, Sanna A, Bosi A et al. Proteomic analysis identifies differentially expressed proteins in AML1/ETO acute myeloid leukemia cells treated with DNMT inhibitors azacitidine and decitabine. Leuk Res 2012; 36: 607–618.
Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 2006; 106: 1794–1803.
Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O'Brien S, Cortes J et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood 2007; 109: 52–57.
Jiang Y, Dunbar A, Gondek LP, Mohan S, Rataul M, O'Keefe C et al. Aberrant DNA methylationis a dominant mechanism in MDS progression to AML. Blood 2009; 113: 1315–1325.
Kulasekararaj AG, Mohamedali AM, Smith AE . Polycomb complex group gene mutations and their prognostic relevance in 5-azacitidine treated myelodysplastic syndrome patient blood. Blood 2010; 116, (abstract 125).
Stresemann C, Lyko F . Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer 2008; 123: 8–13.
Momparler RL, Derse D . Kinetics of phosphorylation of 5-aza-2'-deoxyycytidine by deoxycytidine kinase. Biochem Pharmacol 1979; 28: 1443–1444.
Qin T, Castoro R, El Ahdab S, Jelinek J, Wang X, Si J et al. Mechanisms of resistance to decitabine in the myelodysplastic syndrome. PLoS One 2011; 6: e23372.
Saiki Y, Yoshino Y, Fujimura H, Manabe T, Kudo Y, Shimada M et al. DCK is frequently inactivated in acquired gemcitabine-resistant human cancer cells. Biochem Biophys Res Commun 2012; 421: 98–104.
Van Rompay AR, Norda A, Lindén K, Johansson M, Karlsson A . Phosphorylation of uridine and cytidine nucleoside analogs by two human uridine-cytidine kinases. Mol Pharmacol 2001; 59: 1181–1186.
Lamba JK, Crews K, Pounds S, Schuetz EG, Gresham J, Gandhi V et al. Pharmacogenetics of deoxycytidine kinase: identification and characterization of novel genetic variants. J Pharmacol Exp Ther 2007; 323: 935–945.
Veuger MJ, Honders MW, Landegent JE, Willemze R, Barge RM . High incidence of alternatively spliced forms of deoxycytidine kinase in patients with resistant acute myeloid leukemia. Blood 2000; 96: 1517–1524.
Tattersall MH, Ganeshaguru K, Hoffbrand AV . Mechanisms of resistance of human acute leukaemia cells to cytosine arabinoside. Br J Haematol 1974; 27: 39–46.
Albertioni F, Lindemalm S, Reichelova V, Pettersson B, Eriksson S, Juliusson G et al. Pharmacokinetics of cladribine in plasma and its 5'-monophosphate and 5'-triphosphate in leukemic cells of patients with chronic lymphocytic leukemia. Clin Cancer Res 1998; 4: 653–658.
Acknowledgements
We thank Stacey Rose, PhD, from MediTech Media Ltd for editorial support, and Celgene Corporation. Research support was provided by Regione Toscana, Bando Salute 2009; Ente Cassa di Risparmio di Firenze (ECR); and Ministero per l’Istruzione, l’Università e la Ricerca (MIUR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
VS has received honoraria from Celgene, Janssen and Novartis. MTV received honoraria and support for travelling from Roche, Celgene and Novartis. DC is on the advisory board of Novartis. AB is in the speaker bureau of Novartis, Gilead, BMS, Celgene, Munidpharma, Sanofi and Roche. All the other authors declare no conflict of interest.
Additional information
Author Contributions
VS conceived and designed the study. AV supervised the experiments, performed data analysis and wrote the article. EM, AR and FB performed gene expression, methylation and siRNA experiments. AM and FC did sequence analysis of UCK1. MF and OK performed TET2 mutations. DC, VG and MTV provided patient samples and clinical data. AV, AS and AG were involved in the statistical analysis. AB and VS supervised the study. All authors read the article for the intellectual content, collaborated on the final approval and contributed to the final version to be published.
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Valencia, A., Masala, E., Rossi, A. et al. Expression of nucleoside-metabolizing enzymes in myelodysplastic syndromes and modulation of response to azacitidine. Leukemia 28, 621–628 (2014). https://doi.org/10.1038/leu.2013.330
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2013.330
Keywords
This article is cited by
-
Ubiquitin-specific protease 28: the decipherment of its dual roles in cancer development
Experimental Hematology & Oncology (2023)
-
Therapeutic Targets in Myelodysplastic Neoplasms: Beyond Hypomethylating Agents
Current Hematologic Malignancy Reports (2023)
-
Hypomethylating agents (HMA) for the treatment of acute myeloid leukemia and myelodysplastic syndromes: mechanisms of resistance and novel HMA-based therapies
Leukemia (2021)
-
Decitabine- and 5-azacytidine resistance emerges from adaptive responses of the pyrimidine metabolism network
Leukemia (2021)
-
Clinical, molecular, and prognostic correlates of number, type, and functional localization of TET2 mutations in chronic myelomonocytic leukemia (CMML)—a study of 1084 patients
Leukemia (2020)