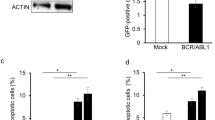
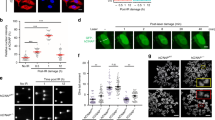
Abstract
Tyrosine kinase inhibitors (TKIs) revolutionized the treatment of chronic myeloid leukemia in chronic phase (CML-CP). Unfortunately, 25% of TKI-naive patients and 50–90% of patients developing TKI-resistance carry CML clones expressing TKI-resistant BCR-ABL1 kinase mutants. We reported that CML-CP leukemia stem and progenitor cell populations accumulate high amounts of reactive oxygen species, which may result in accumulation of uracil derivatives in genomic DNA. Unfaithful and/or inefficient repair of these lesions generates TKI-resistant point mutations in BCR-ABL1 kinase. Using an array of specific substrates and inhibitors/blocking antibodies we found that uracil DNA glycosylase UNG2 were inhibited in BCR-ABL1-transformed cell lines and CD34+ CML cells. The inhibitory effect was not accompanied by downregulation of nuclear expression and/or chromatin association of UNG2. The effect was BCR-ABL1 kinase-specific because several other fusion tyrosine kinases did not reduce UNG2 activity. Using UNG2-specific inhibitor UGI, we found that reduction of UNG2 activity increased the number of uracil derivatives in genomic DNA detected by modified comet assay and facilitated accumulation of ouabain-resistant point mutations in reporter gene Na+/K+ATPase. In conclusion, we postulate that BCR-ABL1 kinase-mediated inhibition of UNG2 contributes to accumulation of point mutations responsible for TKI resistance causing the disease relapse, and perhaps also other point mutations facilitating malignant progression of CML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Melo JV, Barnes DJ . Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer 2007; 7: 441–453.
Marley SB, Gordon MY . Chronic myeloid leukaemia: stem cell derived but progenitor cell driven. Clin Sci 2005; 109: 13–25.
Foroni L, Gerrard G, Nna E, Khorashad JS, Stevens D, Swale B et al. Technical aspects and clinical applications of measuring BCR-ABL1 transcripts number in chronic myeloid leukemia. Am J Hematol 2009; 84: 517–522.
White DL, Hughes TP . Predicting the response of CML patients to tyrosine kinase inhibitor therapy. Curr Hematol Malig Rep 2011; 6: 88–95.
Skorski T . BCR-ABL1 kinase: hunting an elusive target with new weapons. Chem Biol 2011; 18: 1352–1353.
Willis SG, Lange T, Demehri S, Otto S, Crossman L, Niederwieser D et al. High-sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood 2005; 106: 2128–2137.
Gruber FX, Ernst T, Porkka K, Engh RA, Mikkola I, Maier J et al. Dynamics of the emergence of dasatinib and nilotinib resistance in imatinib-resistant CML patients. Leukemia 2012; 26: 172–177.
Perrotti D, Jamieson C, Goldman J, Skorski T . Chronic myeloid leukemia: mechanisms of blastic transformation. J Clin Invest 2010; 120: 2254–2264.
Sorel N, Bonnet ML, Guillier M, Guilhot F, Brizard A, Turhan AG . Evidence of ABL-kinase domain mutations in highly purified primitive stem cell populations of patients with chronic myelogenous leukemia. Biochem Biophys Res Commun 2004; 323: 728–730.
Koptyra M, Cramer K, Slupianek A, Richardson C, Skorski T . BCR/ABL promotes accumulation of chromosomal aberrations induced by oxidative and genotoxic stress. Leukemia 2008; 22: 1969–1972.
Schar P . Spontaneous DNA damage, genome instability, and cancer--when DNA replication escapes control. Cell 2001; 104: 329–332.
Jackson AL, Loeb LA . The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res 2001; 477: 7–21.
Gutteridge JM, Halliwell B . Free radicals and antioxidants in the year 2000. A historical look to the future. Ann NY Acad Sci 2000 899: 136–147.
Lu AL, Li X, Gu Y, Wright PM, Chang DY . Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem Biophys 2001; 35: 141–170.
Feig DI, Sowers LC, Loeb LA . Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc Natl Acad Sci USA 1994; 91: 6609–6613.
Nieborowska-Skorska M, Kopinski PK, Ray R, Hoser G, Ngaba D, Flis S et al. Rac2-MRC-cIII-generated ROS cause genomic instability in chronic myeloid leukemia stem cells and primitive progenitors. Blood 2012; 119: 4253–4263.
Mitra S, Boldogh I, Izumi T, Hazra TK . Complexities of the DNA base excision repair pathway for repair of oxidative DNA damage. Environ Mol Mutagen 2001; 38: 180–190.
Dizdaroglu M, Karakaya A, Jaruga P, Slupphaug G, Krokan HE . Novel activities of human uracil DNA N-glycosylase for cytosine-derived products of oxidative DNA damage. Nucleic Acids Res 1996; 24: 418–422.
Matsubara M, Tanaka T, Terato H, Ohmae E, Izumi S, Katayanagi K et al. Mutational analysis of the damage-recognition and catalytic mechanism of human SMUG1 DNA glycosylase. Nucleic Acids Res 2004; 32: 5291–5302.
Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT et al. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem 2002; 277: 42205–42213.
Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK et al. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem 2002; 277: 30417–30420.
Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M et al. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J Biol Chem 2000; 275: 32422–32429.
Visnes T, Doseth B, Pettersen HS, Hagen L, Sousa MM, Akbari M et al. Uracil in DNA and its processing by different DNA glycosylases. Philos Trans R Soc Lond B Biol Sci 2009; 364: 563–568.
Boiteux S, Radicella JP . Base excision repair of 8-hydroxyguanine protects DNA from endogenous oxidative stress. Biochimie 1999; 81: 59–67.
Stoklosa T, Poplawski T, Koptyra M, Nieborowska-Skorska M, Basak G, Slupianek A et al. BCR/ABL inhibits mismatch repair to protect from apoptosis and induce point mutations. Cancer Res 2008; 68: 2576–2580.
Nieborowska-Skorska M, Hoser G, Kossev P, Wasik MA, Skorski T . Complementary functions of the antiapoptotic protein A1 and serine/threonine kinase pim-1 in the BCR/ABL-mediated leukemogenesis. Blood 2002; 99: 4531–4539.
Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell 2001; 8: 795–806.
Slupianek A, Hoser G, Majsterek I, Bronisz A, Malecki M, Blasiak J et al. Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G(2)/M phase, and protection from apoptosis. Mol Cell Biol 2002; 22: 4189–4201.
Slupianek A, Poplawski T, Jozwiakowski SK, Cramer K, Pytel D, Stoczynska E et al. BCR/ABL stimulates WRN to promote survival and genomic instability. Cancer Res 2011; 71: 842–851.
Dignam JD, Lebovitz RM, Roeder RG . Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 1983; 11: 1475–1489.
Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F et al. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem 2002; 277: 39926–39936.
Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE et al. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell 1995; 80: 869–878.
Blasiak J, Gloc E, Drzewoski J, Wozniak K, Zadrozny M, Skorski T et al. Free radical scavengers can differentially modulate the genotoxicity of amsacrine in normal and cancer cells. Mutat Res 2003; 535: 25–34.
Koptyra M, Falinski R, Nowicki MO, Stoklosa T, Majsterek I, Nieborowska-Skorska M et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood 2006; 108: 319–327.
Sharbeen G, Cook AJ, Lau KK, Raftery J, Yee CW, Jolly CJ . Incorporation of dUTP does not mediate mutation of A:T base pairs in Ig genes in vivo. Nucleic Acids Res 2010; 38: 8120–8130.
Kreutzer DA, Essigmann JM . Oxidized, deaminated cytosines are a source of C --> T transitions in vivo. Proc Natl Acad Sci USA 1998; 95: 3578–3582.
Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T . Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J 2001; 20: 4278–4286.
Eide CA, Adrian LT, Tyner JW, Mac Partlin M, Anderson DJ, Wise SC et al. The ABL switch control inhibitor DCC-2036 is active against the chronic myeloid leukemia mutant BCR-ABLT315I and exhibits a narrow resistance profile. Cancer Res 2011; 71: 3189–3195.
Nowicki MO, Falinski R, Koptyra M, Slupianek A, Stoklosa T, Gloc E et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood 2004; 104: 3746–3753.
Xiao Y, Zou P, Wang J, Song H, Zou J, Liu L . Lower phosphorylation of p38 MAPK blocks the oxidative stress-induced senescence in myeloid leukemic CD34(+)CD38 (−) cells. J Huazhong Univ Sci Technolog Med Sci 2012; 32: 328–333.
Krokan HE, Drablos F, Slupphaug G . Uracil in DNA--occurrence, consequences and repair. Oncogene 2002; 21: 8935–8948.
Branford S, Rudzki Z, Walsh S, Grigg A, Arthur C, Taylor K et al. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood 2002; 99: 3472–3475.
Branford S, Rudzki Z, Walsh S, Parkinson I, Grigg A, Szer J et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis. Blood 2003; 102: 276–283.
Al-Ali HK, Heinrich MC, Lange T, Krahl R, Mueller M, Muller C et al. High incidence of BCR-ABL kinase domain mutations and absence of mutations of the PDGFR and KIT activation loops in CML patients with secondary resistance to imatinib. Hematol J 2004; 5: 55–60.
Willis S, Lange T, Demehri S, Otto S, Crossman L, Niederwieser D et al. High sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood 2005; 24: 24.
Griswold IJ, MacPartlin M, Bumm T, Goss VL, O'Hare T, Lee KA et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol Cell Biol 2006; 26: 6082–6093.
Jiang X, Saw KM, Eaves A, Eaves C . Instability of BCR-ABL gene in primary and cultured chronic myeloid leukemia stem cells. J Natl Cancer Inst 2007; 99: 680–693.
Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 2001; 293: 876–880.
Skorski T . Chronic myeloid leukemia cells refractory/resistant to tyrosine kinase inhibitors are genetically unstable and may cause relapse and malignant progression to the terminal disease state. Leuk Lymphoma 2011; 52 (Suppl 1): 23–29.
Grant H, Jiang X, Stebbing J, Foroni L, Craddock C, Griffiths M et al. Analysis of BCR-ABL1 tyrosine kinase domain mutational spectra in primitive chronic myeloid leukemia cells suggests a unique mutator phenotype. Leukemia 2010; 24: 1817–1821.
Canitrot Y, Capp JP, Puget N, Bieth A, Lopez B, Hoffmann JS et al. DNA polymerase beta overexpression stimulates the Rad51-dependent homologous recombination in mammalian cells. Nucleic Acids Res 2004; 32: 5104–5112.
Canitrot Y, Falinski R, Louat T, Laurent G, Cazaux C, Hoffmann JS et al. p210 BCR/ABL kinase regulates nucleotide excision repair (NER) and resistance to UV radiation. Blood 2003; 102: 2632–2637.
Canitrot Y, Hoffmann JS, Calsou P, Hayakawa H, Salles B, Cazaux C . Nucleotide excision repair DNA synthesis by excess DNA polymerase beta: a potential source of genetic instability in cancer cells. Faseb J 2000; 14: 1765–1774.
Canitrot Y, Lautier D, Laurent G, Frechet M, Ahmed A, Turhan AG et al. Mutator phenotype of BCR–ABL transfected Ba/F3 cell lines and its association with enhanced expression of DNA polymerase beta. Oncogene 1999; 18: 2676–2680.
Acknowledgements
This work was supported by NCI R01 CA123014 from the National Cancer Institute.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Slupianek, A., Falinski, R., Znojek, P. et al. BCR-ABL1 kinase inhibits uracil DNA glycosylase UNG2 to enhance oxidative DNA damage and stimulate genomic instability. Leukemia 27, 629–634 (2013). https://doi.org/10.1038/leu.2012.294
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2012.294
Keywords
This article is cited by
-
The importance of personalized medicine in chronic myeloid leukemia management: a narrative review
Egyptian Journal of Medical Human Genetics (2023)
-
Transcriptional control of CBX5 by the RNA-binding proteins RBMX and RBMXL1 maintains chromatin state in myeloid leukemia
Nature Cancer (2021)
-
Tracking of Normal and Malignant Progenitor Cell Cycle Transit in a Defined Niche
Scientific Reports (2016)
-
Leukemia stem cells: the root of chronic myeloid leukemia
Protein & Cell (2015)
-
Natural course and biology of CML
Annals of Hematology (2015)