Abstract
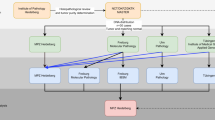
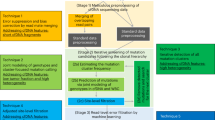
Massively parallel pyrosequencing allows sensitive deep sequencing to detect molecular aberrations. Thus far, data are limited on the technical performance in a clinical diagnostic setting. Here, we investigated as an international consortium the robustness, precision and reproducibility of amplicon next-generation deep sequencing across 10 laboratories in eight countries. In a cohort of 18 chronic myelomonocytic leukemia patients, mutational analyses were performed on TET2, a frequently mutated gene in myeloproliferative neoplasms. Additionally, hotspot regions of CBL and KRAS were investigated. The study was executed using GS FLX sequencing instruments and the small volume 454 Life Sciences Titanium emulsion PCR setup. We report a high concordance in mutation detection across all laboratories, including a robust detection of novel variants, which were undetected by standard Sanger sequencing. The sensitivity to detect low-level variants present with as low as 1–2% frequency, compared with the 20% threshold for Sanger-based sequencing is increased. Together with the output of high-quality long reads and fast run time, we demonstrate the utility of deep sequencing in clinical applications. In conclusion, this multicenter analysis demonstrated that amplicon-based deep sequencing is technically feasible, achieves high concordance across multiple laboratories and allows a broad and in-depth molecular characterization of cancer specimens with high diagnostic sensitivity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Metzker ML . Sequencing technologies—the next generation. Nat Rev Genet 2010; 11: 31–46.
Voelkerding KV, Dames S, Durtschi JD . Next generation sequencing for clinical diagnostics-principles and application to targeted resequencing for hypertrophic cardiomyopathy: a paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn 2010; 12: 539–551.
Thomas RK, Greulich H, Yuza Y, Lee JC, Tengs T, Feng W et al. Detection of oncogenic mutations in the EGFR gene in lung adenocarcinoma with differential sensitivity to EGFR tyrosine kinase inhibitors. Cold Spring Harb Symp Quant Biol 2005; 70: 73–81.
Grossmann V, Kohlmann A, Zenger M, Schindela S, Eder C, Weissmann S et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia 2011; 25: 557–560.
Grossmann V, Kohlmann A, Eder C, Haferlach C, Kern W, Cross NC et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia 2011; 25: 877–879.
Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol 2010; 28: 3858–3865.
Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gaken J, Lea NC et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood 2010; 116: 3923–3932.
Bentley G, Higuchi R, Hoglund B, Goodridge D, Sayer D, Trachtenberg EA et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens 2009; 74: 393–403.
Holcomb CL, Hoglund B, Anderson MW, Blake LA, Bohme I, Egholm M et al. A multi-site study using high-resolution HLA genotyping by next generation sequencing. Tissue Antigens 2011; 77: 206–217.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009; 360: 2289–2301.
Bowen DT, Frew ME, Hills R, Gale RE, Wheatley K, Groves MJ et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood 2005; 106: 2113–2119.
Dunbar AJ, Gondek LP, O’Keefe CL, Makishima H, Rataul MS, Szpurka H et al. 250 K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res 2008; 68: 10349–10357.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437: 376–380.
Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med 2006; 12: 852–855.
Klein HU, Bartenhagen C, Kohlmann A, Grossmann V, Ruckert C, Haferlach T et al. R453Plus1Toolbox: an R/Bioconductor package for analyzing Roche 454 Sequencing data. Bioinformatics 2011; 27: 1162–1163.
Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2011; 39: D38–D51.
Noether GE . Introduction to Statistics: The Nonparametric Way. Springer: New York, 1991.
Wellek S . Testing Statistical Hypotheses of Equivalence. Chapman & Hall/CRC: Boca Raton, 2003.
Bland JM, Altman DG . Measuring agreement in method comparison studies. Stat Methods Med Res 1999; 8: 135–160.
Baccarani M, Pane F, Saglio G . Monitoring treatment of chronic myeloid leukemia. Haematologica 2008; 93: 161–169.
Ernst T, Erben P, Muller MC, Paschka P, Schenk T, Hoffmann J et al. Dynamics of BCR-ABL mutated clones prior to hematologic or cytogenetic resistance to imatinib. Haematologica 2008; 93: 186–192.
Zenz T, Eichhorst B, Busch R, Denzel T, Habe S, Winkler D et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol 2010; 28: 4473–4479.
Kohlmann A, Grossmann V, Schindela S, Kern W, Haferlach C, Haferlach T et al. Ultra-deep next-generation sequencing detects RUNX1 mutations with unprecedented sensitivity and allows to monitor minimal residual disease in 116 samples from MDS and AML patients. Blood 2010; 116: 1691a.
Simen BB, Simons JF, Hullsiek KH, Novak RM, Macarthur RD, Baxter JD et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis 2009; 199: 693–701.
Grossmann V, Schnittger S, Schindela S, Klein HU, Eder C, Dugas M et al. Strategy for robust detection of insertions, deletions, and point mutations in CEBPA, a GC-rich content gene, using 454 next-generation deep-sequencing technology. J Mol Diagn 2011; 13: 129–136.
Acknowledgements
Study supplies and training courses were provided by Roche Diagnostics GmbH Penzberg, Germany. We would like to thank Verena Lutz, Bruno Frey and Wulf Fischer-Knuppertz for their support throughout the conduct of the study. Research in this work was further supported by the European LeukemiaNet (Work Package 13).
Author Contribution
AK and TH designed the study. SW, SB, TC, HC, EH-B, BG, VG, BH, CG, II, JHJ, GtK, LvdL, GM, KM, MRS, BT, PV and BDY performed research and generated data. H-UK, SW, KH, MD and AK analyzed and interpreted the data. AK, H-UK and SW wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
TH is a part-owner of the MLL Munich Leukemia Laboratory GmbH. AK, SW and VG are employed by MLL Munich Leukemia Laboratory GmbH. AK has received honoraria from Roche Diagnostics GmbH Mannheim, Germany. Other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Kohlmann, A., Klein, HU., Weissmann, S. et al. The Interlaboratory RObustness of Next-generation sequencing (IRON) study: a deep sequencing investigation of TET2, CBL and KRAS mutations by an international consortium involving 10 laboratories. Leukemia 25, 1840–1848 (2011). https://doi.org/10.1038/leu.2011.155
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2011.155
Keywords
This article is cited by
-
Sporadic gastric juvenile polyposis with a novel SMAD4 nonsense mutation in a mosaic pattern
Clinical Journal of Gastroenterology (2024)
-
Next-generation sequencing for BCR-ABL1 kinase domain mutation testing in patients with chronic myeloid leukemia: a position paper
Journal of Hematology & Oncology (2019)
-
The role of EIF1AX in thyroid cancer tumourigenesis and progression
Journal of Endocrinological Investigation (2019)
-
Genomic subtyping and therapeutic targeting of acute erythroleukemia
Nature Genetics (2019)
-
Mutations in TP53 and JAK2 are independent prognostic biomarkers in B-cell precursor acute lymphoblastic leukaemia
British Journal of Cancer (2017)