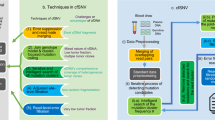
Abstract
Cell-free DNA (cfDNA) in blood, viewed as a surrogate for tumor biopsy, has many clinical applications, including diagnosing cancer, guiding cancer treatment and monitoring treatment response. All these applications depend on an indispensable, yet underdeveloped task: detecting somatic mutations from cfDNA. The task is challenging because of the low tumor fraction in cfDNA. Recently, we developed the computational method cfSNV, the first method that comprehensively considers the properties of cfDNA for the sensitive detection of mutations from cfDNA. cfSNV vastly outperformed the conventional methods that were developed primarily for calling mutations from solid tumor tissues. cfSNV can accurately detect mutations in cfDNA even with medium-coverage (e.g., ≥200×) sequencing, which makes whole-exome sequencing (WES) of cfDNA a viable option for various clinical utilities. Here, we present a user-friendly cfSNV package that exhibits fast computation and convenient user options. We also built a Docker image of it, which is designed to enable researchers and clinicians with a limited computational background to easily carry out analyses on both high-performance computing platforms and local computers. Mutation calling from a standard preprocessed WES dataset (~250× and ~70 million base pair target size) can be carried out in 3 h on a server with eight virtual CPUs and 32 GB of random access memory.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
No new genomic sequencing data were generated in this study. The datasets used in this protocol included (i) the example data, which are available at the European Nucleotide Archive under the accession numbers ERR850376 and ERR852106; and (ii) the test demo data, which are available at https://zenodo.org/record/7191202/files/demo_data.tar.gz.
Code availability
cfSNV can be obtained at https://github.com/jasminezhoulab/cfSNV_docker. It can be freely used for educational and research purposes by nonprofit institutions and U.S. government agencies only under the UCLA Academic Software License. For information on use for a commercial purpose or by a commercial or for-profit entity, please contact Xianghong Jasmine Zhou (XJZhou@mednet.ucla.edu) and Wenyuan Li (WenyuanLi@mednet.ucla.edu).
Change history
03 January 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41596-023-00952-2
References
VanderLaan, P. A. et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer 84, 39–44 (2014).
Murtaza, M. et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 6, 8760 (2015).
Phallen, J. et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9, eaan2415 (2017).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Ueda, M. et al. Somatic mutations in plasma cell-free DNA are diagnostic markers for esophageal squamous cell carcinoma recurrence. Oncotarget 7, 62280–62291 (2016).
Adalsteinsson, V. A. et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017).
Camus, V. et al. Digital PCR for quantification of recurrent and potentially actionable somatic mutations in circulating free DNA from patients with diffuse large B-cell lymphoma. Leuk. Lymphoma 57, 2171–2179 (2016).
Rothwell, D. G. et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat. Med. 25, 738–743 (2019).
Li, S. et al. Sensitive detection of tumor mutations from blood and its application to immunotherapy prognosis. Nat. Commun. 12, 1–14 (2021).
Goldberg, S. B. et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 24, 1872–1880 (2018).
Iwama, E. et al. Monitoring of somatic mutations in circulating cell-free DNA by digital PCR and next-generation sequencing during afatinib treatment in patients with lung adenocarcinoma positive for EGFR activating mutations. Ann. Oncol. 28, 136–141 (2017).
Fontanilles, M. et al. Non-invasive detection of somatic mutations using next-generation sequencing in primary central nervous system lymphoma. Oncotarget 8, 48157–48168 (2017).
Chaudhuri, A. A. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 7, 1394–1403 (2017).
Li, S. et al. cfTrack, a method of exome-wide mutation analysis of cell-free DNA to simultaneously monitor the full spectrum of cancer treatment outcomes including MRD, recurrence, and evolution. Clin. Cancer Res. 28, 1841–1853 (2022).
Choudhury, A. D. et al. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight 3, e122109 (2018).
Li, S. et al. cfSNV: a software tool for the sensitive detection of somatic mutations from cell-free DNA. Jasminezhoulab/cfSNV_docker: cfSNV docker image. Available at https://github.com/jasminezhoulab/cfSNV_docker (2022).
Jiang, P. et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl Acad. Sci. USA 112, E1317–E1325 (2015).
Jiang, P. et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc. Natl Acad. Sci. USA 115, E10925–E10933 (2018).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–461 (2017).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Van der Auwera, G. A. et al. From FastQ data to high‐confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinforma. 43, 11.10.1–11.10.33 (2013).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Pellini, B. & Chaudhuri, A. A. Circulating tumor DNA minimal residual disease detection of non–small-cell lung cancer treated with curative intent. J. Clin. Oncol. 40, 567–575 (2022).
Roth, A. et al. JointSNVMix: a probabilistic model for accurate detection of somatic mutations in normal/tumour paired next-generation sequencing data. Bioinformatics 28, 907–913 (2012).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Kothen-Hill, S. T. et al. Deep learning mutation prediction enables early stage lung cancer detection in liquid biopsy. Available at https://openreview.net/forum?id=H1DkN7ZCZ (2018).
Zviran, A. et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 26, 1114–1124 (2020).
Koboldt, D. C. Best practices for variant calling in clinical sequencing. Genome Med. 12, 1–13 (2020).
Chen, Z. et al. Systematic comparison of somatic variant calling performance among different sequencing depth and mutation frequency. Sci. Rep. 10, 3501 (2020).
Xu, C. et al. A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput. Struct. Biotechnol. J. 16, 15–24 (2018).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Broad Institute. Picard tools. Available at https://broadinstitute.github.io/picard/ (2019).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Mose, L. E. et al. ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics 30, 2813–2815 (2014).
Opasic, L. et al. How many samples are needed to infer truly clonal mutations from heterogenous tumours? BMC Cancer 19, 1–11 (2019).
Butler, T. M. et al. Exome sequencing of cell-free DNA from metastatic cancer patients identifies clinically actionable mutations distinct from primary disease. PloS One 10, e0136407 (2015).
Kurtz, D. M. et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat. Biotechnol. 39, 1537–1547 (2021).
Liebs, S. et al. Liquid biopsy assessment of synchronous malignancies: a case report and review of the literature. ESMO Open 4, e000528 (2019).
Ramesh, N. et al. Decoding the evolutionary response to prostate cancer therapy by plasma genome sequencing. Genome Biol. 21, 1–22 (2020).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Merkel, D. Docker: lightweight linux containers for consistent development and deployment. Linux J. 2014, 2 (2014).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Arnold, K., Gosling, J. & Holmes, D. The Java Programming Language (Addison Wesley Professional, 2005).
Van Rossum, G. & Drake, F. L. Python 3 Reference Manual (CreateSpace, Scotts Valley, CA, 2009).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
McKinney, W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference, Vol. 445 (SCIPY, 2010).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
R Core Team. R: a language and environment for statistical computing. Available at https://www.R-project.org/ (2018).
Eddelbuettel, D. & Romain, F. Rcpp: seamless R and C++ integration. J. Stat. Softw. 40, 1–18 (2011).
Sherry, S. T., Ward, M. & Sirotkin, K. dbSNP—database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 9, 677–679 (1999).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Acknowledgements
This work was supported by National Cancer Institute grant nos. U01CA230705 and R01CA264864 to X.J.Z., R01CA246329 to X.J.Z. and W.L. and U01CA237711 to W.L.
Author information
Authors and Affiliations
Contributions
S.L., R.H., C.S., T.-Y.K. and C.-C.L. developed the protocol. S.L. and R.H. wrote the manuscript. W.L. and X.J.Z. supervised the study. All authors discussed and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
X.J.Z. and W.L. are co-founders of EarlyDiagnostics Inc. X.J.Z. serves on the Board of Directors and has an executive leadership position at EarlyDiagnostics. C.-C.L. and T.-Y.K. are employees of EarlyDiagnostics. X.J.Z. and W.L. are stockholders of EarlyDiagnostics. S.L., C.-C.L. have stock options with EarlyDiagnostics. W.L. is a consultant for EarlyDiagnostics. X.J.Z., S.L. and W.L. are inventors on a patent application submitted by the Regents of the University of California and licensed to EarlyDiagnostics (patent no. US20210125683A1). The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Navonil De Sarkar, Alain Thierry and Zhidong Tu for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key papers using this protocol
Li, S. et al. Nat. Commun. 12, 4172 (2021): https://doi.org/10.1038/s41467-021-24457-2
Li, S. et al. Clin. Cancer Res. 28, 1841–1853 (2022): https://doi.org/10.1158/1078-0432.CCR-21-1242
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, S., Hu, R., Small, C. et al. cfSNV: a software tool for the sensitive detection of somatic mutations from cell-free DNA. Nat Protoc 18, 1563–1583 (2023). https://doi.org/10.1038/s41596-023-00807-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00807-w
This article is cited by
-
cfSNV: a software tool for the sensitive detection of somatic mutations from cell-free DNA
Nature Protocols (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.