Abstract
Maspin (mammary serine protease inhibitor or SerpinB5) acts as a tumor suppressor when overexpressed in aggressive cancer cell lines. However, its role in human cancer is controversial. Maspin expression has been associated with a poor prognosis in some studies, whereas in others, with favorable outcome. The clinical data suggest, however, that nuclear-localized maspin is associated with improved survival. We hypothesized that the tumor suppressor activity of maspin may require nuclear localization, and that the discordance between clinical and experimental reports is a consequence of the variable subcellular distribution of maspin. Furthermore, we surmized that nuclear maspin could function as a tumor suppressor through the regulation of genes involved in tumor growth and invasion. Maspin or maspin fused to a nuclear export signal were expressed in metastatic human breast and epidermoid carcinoma cell lines. We found that pan-cellular localized maspin inhibited in vivo tumor growth and metastasis when assessed in xenograft chicken embryo and murine mammary fat pad injection models. However, when maspin was excluded from the nucleus via a nuclear exclusion signal, it no longer functioned as a metastasis suppressor. Using chromatin immunoprecipitation, we show that nuclear maspin was enriched at the promoter of colony-stimulating factor-1 (CSF-1) and associated with diminished levels of CSF-1 mRNA. Our findings demonstrate that the nuclear localization of maspin is required for its tumor and metastasis suppressor functions in vivo, and suggest that its mechanism of action involves, in part, direct association of maspin with target genes.
Similar content being viewed by others
Main
Maspin (mammary serine protease inhibitor or SerpinB5), a member of the serpin family of serine protease inhibitors, was identified in 1994 as a tumor and metastasis suppressor.1 Overexpression of maspin inhibits cell motility and invasion in vitro, and decreases tumor formation, metastasis and angiogenesis in vivo.2 Initially, secreted maspin was thought to inhibit migration and increase cell adhesion via interaction with β1-integrin and other extracellular matrix components.2 However, the molecular mechanism of such activity has yet to be convincingly demonstrated, and remains the subject of debate. Indeed, the very existence of extracellular maspin has recently been questioned.3 Maspin has also been detected in the nucleus.2
Maspin expression has been characterized in different cancers, particularly in breast cancer. Several studies showed that maspin mRNA is reduced in primary tumors and undetectable in metastases.4, 5 Conversely, others have shown that increased maspin is associated with poor prognosis.6, 7 These conflicting observations might be explained by distinct subcellular localization of maspin in cancer cells. Indeed, recent studies indicate that nuclear maspin associates with well-differentiated phenotype and improved survival, whereas cytoplasmic maspin is associated with poor prognosis in breast, ovarian and head and neck cancers.8, 9, 10 In breast cancer, cytoplasmic localization of maspin associates with estrogen receptor (ER)-negative tumors and early tumor relapse.8, 11 Clinical data thus suggest an association between nuclear maspin and improved prognosis. Contrary to what has been observed in breast, ovarian and head and neck cancers, various groups have reported an association of nuclear expression of maspin with aggressive behavior in colorectal cancer. It is plausible that the tumor suppressive function of nuclear maspin is dependent upon the organ in which it is expressed, as tissue-specific oncogenic and tumor suppressor activities have been described for other proteins such as Notch.12
To address the apparent discordance between experimental and clinical observations in breast and head and neck cancers, we analyzed the role of maspin subcellular localization on tumor growth and metastasis by expressing maspin or nuclear-excluded maspin in two aggressive cell lines that lack endogenous maspin expression. Our results indicate that nuclear-excluded maspin does not inhibit metastasis. We further show that maspin is associated with chromatin and is recruited to the promoter of the gene encoding the cytokine colony-stimulating factor-1 (CSF-1).
MATERIALS AND METHODS
Cell Culture and Antibodies
GFP–HEp3 cells were obtained from Andrew Zijlstra (San Diego, CA).13 MDA-MB-231-D3H2-LNluc cells (herein called D3H2-LN) were from Caliper LifeSciences (Alameda, CA).14 Stable cell lines were generated by retroviral transduction, and pools of clones were utilized for all experiments. The antibodies used include anti-maspin (G167-70, BD Pharmingen), anti-HA-3F10 (Roche), anti-actin (Sigma-Aldrich), anti-Flag-M2 (Invitrogen), anti-tubulin (Invitrogen) and anti-CSF1 (Santa Cruz Biotechnology).
Plasmid Construction
Oligonucleotides containing Flag and hemagglutinin (HA) tags in tandem were ligated into pcDNA3, as described.15 Human maspin was cloned by PCR amplification from pRSET-maspin (gift from James Whisstock, Monash University). The strong nuclear export sequence (NES) of PKIα (LALKLAGLDI) was inserted in-frame between the carboxyl terminus of maspin and the tags.16 Constructs were then cloned into pBabe using the In-Fusion Advantage PCR cloning Kit (Clonetech).
Spontaneous Metastasis Assays in the Chicken Embryo
These assays were performed, as described in Zijlstra et al,13 using GFP–HEp3 cells stably expressing empty vector or a maspin construct (wild-type maspin, maspin–Flag–HA or NES–Flag–HA). Briefly, 7.5 × 105 cells were placed on the chorioallantoic membrane (CAM) of a 10-day-old chicken embryo. After 7 days of incubation, the lungs of the chickens were harvested and genomic DNA extracted. Quantitative real-time PCR using primers against human Alu sequences or chicken GAPDH was used to detect the presence of human tumor cells in the chick lungs.
Spontaneous Metastasis Assay in the Mouse
Female nude beige mice aged 8–10 weeks (Charles River Laboratories) were injected with D3H2-LN-derived cell lines (2 × 106 cells) in 50% PBS/50% Matrigel (BD Biosciences) into the abdominal mammary fat pad. Tumor growth was monitored weekly for 9 weeks by caliper measurements by a technician blinded to the experimental groups.
RT-PCR
RNA was extracted from flash-frozen primary tumors using the RNeasy Mini kit (Qiagen). cDNA was prepared using Superscript II (Invitrogen) and Q-PCR performed using the LightCycle 480 (Roche).
Indirect Immunofluorescence
Cells were plated on a glass coverslip for 24–48 h. They were stained with anti-HA antibodies (1:500) for 1 h. Nuclei were counterstained with Hoechst (1:50 000). Confocal analysis was performed on a Fluoview 1000 laser scanning confocal microscope (Olympus Canada) using the × 60 Plan Apochromat 1.42 oil objective.
Histology
Tumors, lymph nodes and lungs were paraffin embedded, sectioned and stained with H&E. For immunohistochemistry, sections were incubated with anti-HA (1:200) or anti-maspin antibodies (1:200) for 1 h, followed by a biotinylated polyclonal rabbit anti-rat (1/500, DakoCytomation). Sections were then treated with streptavidin peroxidase and stained with the DAB chromogen.
Chromatin Immunoprecipitation Assay
D3H2-LN–maspin–Flag–HA cells were crosslinked for 10 min with 1% formaldehyde, followed by two washes with cold 1X PBS. Cell lysis buffer contained 50 mM Tris-Hcl (pH 8.0), 10 mM EDTA, 1% SDS supplemented with protease and phosphatase inhibitors (Roche) and 1 μM trichostatin A. Cell lysates were sonicated 6 × for 8 s at power 10. An undiluted aliquot (10%) was reverse crosslinked and used as input. Sonicated chromatin was diluted 8 × in chromatin dilution buffer, containing 10 mM Tris-HCL (pH7.5), 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% deoxycholate, with inhibitors. Immunoprecipitations were performed using anti-Flag antibodies (Sigma F1804) or mouse IgGs (Santa Cruz Biotechnology) with anti-mouse IgG magnetic beads (Dynabeads, Invitrogen) rotating for 3 h at 4°C. Quantification of chromatin immunoprecipitation (ChIP) enrichment by quantitative PCR was done on eluted, purified DNA using the LightCycle 480 (Roche). Primers used for ChIP on the CSF-1 promoter were: CSF-1: 5′-CTGCTCTCGGTCCGTTTTCTGCTAAG-3′ and 5′-CCAGGCTGATTCAGTGCTAGCGCTCT-3′, and the control region: 5′-TCACACAGTTCCACCCGCTCCAGAAA-3′ and 5′-GGTGTGGTATTGACAAGCCCTCAGGT-3′. The fold enrichment was determined by the percentage of chromatin immunoprecipitated with the antibody related to the IgG immunoprecipitation.
RESULTS
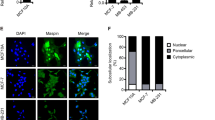
To assess the effects of maspin on tumorigenesis and metastasis, two human cancer cell lines were tested using two well-established in vivo assays. D3H2-LN is a breast cancer cell line that metastasizes to lymph nodes and lungs after mammary fat pad injections in nude mice.14 HEp3 is a head and neck squamous-cell carcinoma cell line that is highly invasive in the chicken embryo.13 Endogenous maspin was not detected in either of the cell lines (Supplementary Figure S1). To unambiguously confirm subcellular localization, we generated a maspin construct that contains in tandem Flag and HA tags at its carboxyl terminus (maspin–Flag–HA, Supplementary Figure S1). Invasion of both cell lines through matrigel in a Boyden chamber assay was reduced by either wild-type maspin or maspin–Flag–HA (Supplementary Figure S1). This result, consistent with previous reports,1, 2 shows that the Flag–HA tag does not interfere with known activity of maspin. We next fused a strong nuclear export signal to maspin to exclude it from the nucleus (NES–Flag–HA, Figure 1a and b). NES–Flag–HA was expressed to levels comparable to maspin–Flag–HA (Figure 1a) and was found to be strictly cytoplasmic (Figure 1b).
Nuclear-excluded maspin does not inhibit metastasis in the chicken embryo. (a) Western blot analysis of total cell lysates from D3H2-LN cells or HEp3 cells stably expressing the indicated constructs. (b) Indirect immunofluorescence using anti-HA antibodies (bottom panel). Nuclei were counterstained with Hoechst (top panel). Magnification × 60. (c) Quantitative analysis of spontaneous metastasis in the chicken embryo. Expression of human Alu relative to chicken GAPDH (chGAPDH) was normalized to the control cells. (d) Quantitative analysis as in c. Expression of huAlu relative to chGAPDH was normalized to the maspin–Flag–HA cells. *P<0.01, Student's t-test.
To assess metastatic ability, GFP–HEp3-derived stable cell lines were inoculated on the CAM of 10-day-old chicken embryos. As expected, wild-type maspin and maspin–Flag–HA significantly reduced lung metastasis formation compared with vector control cells (Figure 1c). In sharp contrast, NES–Flag–HA cells, which express maspin that is excluded from the nucleus, showed no inhibition by maspin (Figure 1d). These results demonstrate that exclusion of maspin from the nucleus prevents it from functioning as a metastasis inhibitor in vivo.
To further characterize in vivo metastatic behavior, maspin–Flag–HA, NES–Flag–HA or vector control D3H2-LN cells were injected into the fourth mammary fat pad of nude beige mice. All cell lines formed tumors in 100% of inoculated animals (Table 1, Figure 2a). Maspin–Flag–HA cells generated significantly smaller primary tumors than vector control or NES–Flag–HA cells. Similar levels of maspin were observed in maspin–Flag–HA and NES–Flag–HA tumors, as shown by RT-PCR (Figure 2b) and western blot (Figure 2c). Immunohistochemistry using anti-HA antibodies further validated the expression and localization patterns of maspin in the primary tumors (Figure 2d). Maspin–Flag–HA transfectants express both nuclear and cytoplasmic maspin, whereas in NES maspin–Flag–HA transfectants, only cytoplasmic maspin is detected. Overall, only minimal differences in HA staining were observed between the invasive front and the tumor centre (Figure 2d, bottom left panel). Interestingly, maspin–Flag–HA transfectants with lymph node metastases tended to show variable cytoplasmic staining and weak-to-absent nuclear staining in both primary tumor and metastases, suggesting loss of nuclear maspin with metastatic competency (Figure 2d, bottom panels).
Nuclear maspin is required for tumor- and metastasis-suppressing effects in mice. (a) Tumor growth of D2H3-LN cells injected into the mammary fat pad. (b) Real-time Q-PCR showing maspin expression relative to 18S rRNA in primary tumors. (c) Western blotting of total extracts isolated from primary tumors. (d) H&E staining and immunohistochemistry of primary tumors and an animal-matched pair of primary tumor and lymph node metastasis. Insets at higher magnification show nuclear staining. Arrowheads denote nucleus. Scale bar=25 μm (10 μm for inset).
Histopathological analysis revealed that vector control cells had an incidence of metastasis of 75% (Table 1). This number dropped to 40% with maspin–Flag–HA-expressing tumors. Metastases derived from control cells exhibited large foci, whereas metastases from maspin–Flag–HA cells were generally microscopic. Interestingly, however, NES–Flag–HA tumors were more aggressive than controls and produced large metastases in 100% of mice. Compared with control mice, NES–Flag–HA produced 10 times more lung metastases, although mean primary tumor sizes were similar (Table 1). Lymph node involvement was the same for vector control and NES–Flag–HA mice, suggesting that nuclear exclusion of maspin resulted in enhanced dissemination via the hematogenous route.
Microarray and proteomic analysis of breast cancer cells following expression of maspin previously revealed variations in gene and protein expression.2, 17 By RT-PCR, we found that the expression of colony-stimulation factor-1 (CSF-1), an important cytokine during metastasis, was altered. Maspin–Flag–HA tumors had a significant reduction in CSF1 mRNA levels compared with vector control and NES–Flag–HA tumors (Figure 3a). Similarly, CSF-1 protein expression was decreased in maspin–Flag–HA tumors compared with vector control and NES–Flag–HA tumors (Figure 3b).
CSF1 is downregulated in maspin–Flag–HA tumors, and maspin binds to the CSF1 promoter. (a) Real-time Q-PCR showing CSF-1 expression relative to 18S rRNA in primary tumors. **, *P<0.05, one-way ANOVA. (b) Western blotting of total extracts isolated from primary tumors. (c) Immunoprecipitation of maspin using Flag antibodies or IgGs from maspin–Flag–HA cells after crosslinking (FA) and sonication. Maspin was detected by western blot (arrow); *, heavy chain. (d) ChIP using Flag antibodies in maspin–Flag–HA cells, with primers specific for the CSF1 promoter or a negative control region. Data presented as normalized fold enrichment against IgGs. *, P=0.005, Student's t-test.
The recently published CSF-1 response signature bears resemblance to breast cancers that have lost nuclear maspin expression; they correlate with high-grade, ER- and progesterone receptor-negative tumors.8, 18 Interestingly, these tumors also associate with increased levels of the orphan nuclear receptor ERRα (NR3B1 and ESRRA), a transcription factor associated with poor prognosis.19, 20 We therefore analyzed ESRRA expression and found reduced mRNA levels in maspin–Flag–HA tumors (Supplementary Figure S2).
To investigate whether nuclear maspin could occupy the CSF1 promoter in vivo and be involved in its regulation, we performed ChIP. We first confirmed that the tagged maspin was specifically immunoprecipitated from chromatin using anti-Flag antibodies (Figure 3c, lane 5). Nothing was immunoprecipitated by IgGs in maspin–Flag–HA cells (Figure 3c, lane 4) or by anti-Flag antibodies in the control cells (Figure 3c, lane 1). Using these same immunoprecipitation conditions, we observed significant enrichment of maspin on the CSF1 promoter (Figure 3d). Importantly, no enrichment was detected on an unrelated control region. Similarly, maspin also bound to the ESRRA promoter, confirming that binding of maspin to promoter regions of genes is a bona fide feature of its mechanism of action (Supplementary Figure S2).
DISCUSSION
We have demonstrated that nuclear localization of maspin is required for its tumor and metastasis suppressor function in vivo. Importantly, our data provide a mechanism for the discordance in maspin expression seen in human cancers, in which maspin has been associated with both good and poor prognosis.4, 6 Our findings also explain the failure of previous experimental studies to identify a tumorigenic activity associated with maspin.1, 2 Finally, we provide a working model to study the specific roles of nuclear and cytoplasmic maspin in cancer.
We found that maspin was undetectable in two invasive and highly metastatic human cancer cell lines, D3H2LN breast carcinoma and HEp3 epithelial carcinoma. Reintroduction of maspin in these two aggressive cell lines prevented them from forming metastases in two distinct in vivo models. These observations are consistent with published reports using other cell lines.1 However, we clearly demonstrate that when maspin was excluded from the nucleus, metastasis inhibition was lost. In fact, tumor cells expressing nucleus-excluded maspin were more metastatic than control cells. Moreover, we report that maspin can bind to the promoter region of CSF1, and CSF1 expression was significantly reduced in tumors expressing maspin–Flag–HA. These data not only suggest that nuclear localization is vital to the tumor suppressive activity of maspin but also further suggest that cytoplasmic and/or secreted maspin does not contribute to this activity.
The mechanism regulating the nuclear localization or exclusion of maspin remains unknown. Examination of the protein sequence did not reveal any homology to previously identified nuclear localization sequences.21 We therefore speculate that maspin either contains a non-classical nuclear localization sequence that is yet to be identified or associates with a partner protein to transit into the nucleus.
To begin to address the mechanism by which nuclear maspin may function as a tumor and metastasis suppressor, we examined several potential maspin-regulated genes. Our data suggests that nuclear maspin could, in part, mediate a cross-talk between the tumor cells and components of the tumor microenvironment through the regulation of gene expression, as we have demonstrated with CSF1. The involvement of CSF-1 in breast cancer progression and metastasis is well known,18 and our data indicate a direct association between its decreased expression and binding of nuclear maspin to its promoter. We have also shown that maspin binds the ESRRA promoter, and that the mRNA levels encoding ERRα were downregulated in maspin–Flag–HA tumors. Thus, at least two genes crucial to progression of breast cancer are negatively regulated by nuclear maspin, supporting a nuclear role for its tumor suppressive activity.
Molecularly, there is also evidence that maspin interacts with and regulates the activity of HDAC1.22 However, in contrast to what Li et al22 published, we saw no increase in acetylated histone H3 in D3H2-LN-maspin-expressing cells (data not shown). We propose that rather than regulating global chromatin acetylation, maspin may affect HDAC1 activity in a gene-specific manner. The idea of serpins as regulators of chromatin is not novel. For example, MENT (myeloid and erythroid nuclear termination stage-specific protein) is a serpin known to participate in chromatin condensation.23 It would therefore be interesting to understand how maspin gets recruited to chromatin and regulates gene expression. In addition, the use of ChIP-on-chip or ChIP-seq technologies would unveil the genes and/or genomic regions bound and regulated by nuclear maspin and give us a broader understanding of its role as a tumor and metastatic suppressor.
Our results show that changes in the cellular localization of maspin can strongly influence whether a cancer is more or less aggressive, without a change in RNA levels. Whereas gene signatures have become essential in our understanding of cancer progression, it is the expression, modification and localization of proteins that are ultimately responsible for the behavior of cancer cells and patient outcomes. There is recent evidence that forcing the reexpression of maspin in maspin-negative tumors using artificial transcription factors is a promising strategy for cancer treatment.24 In terms of subcellular localization, it will be crucial to identify the regulatory pathways mediating the nuclear versus cytoplasmic localization of maspin, as these could offer novel cancer therapeutic targets.
References
Zou Z, Anisowicz A, Hendrix MJ, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science 1994;263:526–529.
Bailey CM, Khalkhali-Ellis Z, Seftor EA, et al. Biological functions of maspin. J Cell Physiol 2006;209:617–624.
Teoh SS, Whisstock JC, Bird PI . Maspin (SERPINB5) is an obligate intracellular serpin. J Biol Chem 2010;285:10862–10869.
Maass N, Hojo T, Rosel F, et al. Down regulation of the tumor suppressor gene maspin in breast carcinoma is associated with a higher risk of distant metastasis. Clin Biochem 2001;34:303–307.
Vecchi M, Confalonieri S, Nuciforo P, et al. Breast cancer metastases are molecularly distinct from their primary tumors. Oncogene 2007;27:2148–2158.
Umekita Y, Ohi Y, Sagara Y, et al. Expression of maspin predicts poor prognosis in breast-cancer patients. Int J Cancer 2002;100:452–455.
Kim DH, Yoon DS, Dooley WC, et al. Association of maspin expression with the high histological grade and lymphocyte-rich stroma in early-stage breast cancer. Histopathology 2003;42:37–42.
Mohsin SK, Zhang M, Clark GM, et al. Maspin expression in invasive breast cancer: association with other prognostic factors. J Pathol 2003;199:432–435.
Sood AK, Fletcher MS, Gruman LM, et al. The paradoxical expression of maspin in ovarian carcinoma. Clin Cancer Res 2002;8:2924–2932.
Marioni G, Staffieri A, Blandamura S . Maspin expression in head and neck carcinoma: subcellular localization matters. J Oral Pathol Med 2010;39:279–280.
Joensuu KM, Leidenius MH, Andersson LC, et al. High expression of maspin is associated with early tumor relapse in breast cancer. Hum Pathol 2009;40:1143–1151.
Roy M, Pear WS, Aster JC . The multifaceted role of Notch in cancer. Curr Opin Genet Dev 2007;17:52–59.
Zijlstra A, Mellor R, Panzarella G, et al. A quantitative analysis of rate-limiting steps in the metastatic cascade using human-specific real-time polymerase chain reaction. Cancer Res 2002;62:7083–7092.
Jenkins DE, Hornig YS, Oei Y, et al. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivo detection of mammary tumors and multiple metastases in immune deficient mice. Breast Cancer Res 2005;7:R444–R454.
DeMasi J, Huh KW, Nakatani Y, et al. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc Natl Acad Sci USA 2005;102:11486–11491.
Henderson BR, Eleftheriou A . A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res 2000;256:213–224.
Chen EI, Florens L, Axelrod FT, et al. Maspin alters the carcinoma proteome. Faseb J 2005;19:1123–1124.
Beck AH, Espinosa I, Edris B, et al. The macrophage colony-stimulating factor 1 response signature in breast carcinoma. Clin Cancer Res 2009;15:778–787.
Suzuki T, Miki Y, Moriya T, et al. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res 2004;64:4670–4676.
Deblois G, Hall JA, Perry MC, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor alpha as a determinant of breast cancer heterogeneity. Cancer Res 2009;69:6149–6157.
Nair R, Carter P, Rost B . NLSdb: database of nuclear localization signals. Nucleic Acids Res 2003;31:397–399.
Li X, Yin S, Meng Y, et al. Endogenous inhibition of histone deacetylase 1 by tumor-suppressive maspin. Cancer Res 2006;66:9323–9329.
McGowan S, Buckle AM, Irving JA, et al. X-ray crystal structure of MENT: evidence for functional loop-sheet polymers in chromatin condensation. EMBO J 2006;25:3144–3155.
Beltran AS, Blancafort P . Reactivation of MASPIN in non-small cell lung carcinoma (NSCLC) cells by artificial transcription factors (ATFs). Epigenetics 2011;6:229–235.
Acknowledgements
We greatly appreciated the experience of Ms Geneviève Deblois in ChIP. The CSF-1 antibody was a generous gift from Dr WJ Muller and the trichostatin A from Dr XJ Yang. This research was supported by a Postdoctoral Fellowship Award from the Terry Fox Foundation (B Goulet), Grant no. 016506 from Canadian Breast Cancer Research Alliance (ABT, AFC, JDL), Grant no. 018176 from Canadian Cancer Society Research Institute (JDL) and Grant no. MOP-64275 from Canadian Institutes of Health Research (VG). AFC holds a Canada Research Chair in Oncology, supported by the Canada Research Chairs Program.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
The role of maspin localization in tumor growth and metastasis is revealed by expressing maspin or nuclear-excluded maspin in cells that lack endogenous maspin. Exclusion of maspin from the nucleus abolishes metastasis suppression; only the nuclear form is associated with diminished levels of colony-stimulating factor-1, a cytokine involved in tumor growth and invasion.
Supplementary information
Rights and permissions
About this article
Cite this article
Goulet, B., Kennette, W., Ablack, A. et al. Nuclear localization of maspin is essential for its inhibition of tumor growth and metastasis. Lab Invest 91, 1181–1187 (2011). https://doi.org/10.1038/labinvest.2011.66
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2011.66
Keywords
This article is cited by
-
PI3K-AKT, JAK2-STAT3 pathways and cell–cell contact regulate maspin subcellular localization
Cell Communication and Signaling (2021)
-
Role of cytoplasmic localization of maspin in promoting cell invasion in breast cancer with aggressive phenotype
Scientific Reports (2021)
-
TRIM21–SERPINB5 aids GMPS repression to protect nasopharyngeal carcinoma cells from radiation-induced apoptosis
Journal of Biomedical Science (2020)
-
Quantification of cancer cell extravasation in vivo
Nature Protocols (2016)
-
Nuclear location of tumor suppressor protein maspin inhibits proliferation of breast cancer cells without affecting proliferation of normal epithelial cells
BMC Cancer (2014)