Abstract
The south of Tunisia is characterized by marked ethnic diversity, highlighted by the coexistence of native Berbers with Blacks, Jews and Arab-speaking populations. Despite this heterogeneity, genetic anthropology studies investigating the origin of current Southern Tunisians were rarely reported. We examined human leukocyte antigen (HLA) class I (A, B) and class II (DRB1, DQB1) gene profiles of 250 unrelated Southern Tunisians, and compared them with those of Arab-speaking communities, along with Mediterranean and sub-Sahara African populations using genetic distances, neighbor-joining dendrograms, correspondence and haplotype analysis. In total, 137 HLA alleles were detected, which comprised 32 HLA-A, 52 HLA-B, 32 DRB1 and 21 DQB1 alleles. The most frequent alleles were HLA-A*02:01(18.02%), HLA-B*50:01 (9.11%), HLA-DRB1*07:01 (22.06%) and HLA-DQB1*02:01 (17.21%). All pairs of HLA loci show significant linkage disequilibrium. The four loci depict negative Fnd (the normalized deviate of the homozygosity) values indicating an overall trend to balancing selection. Southern Tunisians appear to be closely related to others Tunisian populations including Berbers, North Africans and Iberians. On the contrary, Southern Tunisians were distinct from Palestinian, Lebanese and Jordanian Middle Eastern Arab-speaking population, despite the deep Arab incursions and Arabization that affected Southern Tunisia. In addition, Southern Tunisians were distant from many sub-Saharan communities, evidenced by genetic distance analysis. Collectively, this indicates a limited genetic contribution of Arab invasion and Black caravans on the makeup of Southern Tunisian gene pool.
Similar content being viewed by others
Introduction
The human leukocyte antigen (HLA) region is the most polymorphic region in the human genome, and extends ~3.6 Mb on the short arm of chromosome 6,1 with more than 13 000 HLA alleles deposited in the IMGT/HLA database.2 The high polymorphism, tight linkage, the random association of alleles and the perpetuation of allelic lineages over time make HLA genetic markers an invaluable tool in unraveling the human past. The crucial information relating to amount, pattern and distribution of genetic variation of HLA genetic markers in different populations enable us to correlate genetic profile of populations and their past migrations in the determination of their origin.3
Present-day Tunisians derive their origins from native Berbers and civilizations, which either invaded or migrated to Tunisia throughout history. These included Phoenicians (ancestors of Lebanese) and Romans, who were succeeded by the invasion of European tribes, such as the Vandals.4 In the 7th century ad, Muslim people coming from the Arabian Peninsula and the Middle East invaded Tunisia, and founded the first Islamic city in North Africa, Kairouan. A significant admixture of the Tunisian population was with the Islamic invasion of the South of Tunisia in 11th century ad by Arabian Peninsula tribes.5 Tunisia was later invaded by Turks (Ottoman) and Europeans, and became a French protectorate until the formal independence from France was obtained in 1956.
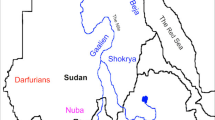
Southern Tunisia (Figure 1) is characterized by high ethnical diversity, and its present population (~12% of the total population according to official census in 2014) comprises Berbers, Blacks, Jews and Arab-speaking populations. The native Berbers reside in geographically isolated communities (Matmata, Djerba, Douiret and so on), and speak Shleuh and Arabic. Tunisian Blacks are more frequent in Southern (particularly in Douz, Kebili and Tataouine) than Northern Tunisia.6 Tunisian Jews (estimated at 1500; <0.1% of total population) cluster in Djerba Island, and they probably came from Andalus and Levant.7 In addition, a large part of Southern Tunisians think that their origin, culture and religion came from Banu Hilal and Banu Soulaym tribes who invaded Southern Tunisians in the eleventh century.8 On the other hand, some studies claim that Southern Tunisians are indeed (native) Berbers, who were ‘Arabized’ during the Arab invasions in the eleventh century.9, 10 Our study is an attempt to understand the most likely origin of this population, which was considered the gateway for Arabs to invade all the Maghreb.
To shed some light on the origin of Southern Tunisians, we investigate the distribution of HLA class I and class II alleles among Southern Tunisians and compared them with those of other Tunisians, Middle Eastern Arab-speaking, Mediterranean and sub-Saharan populations (Table 1). This comparison is instrumental toward a better understanding of the origin and culture of Southern Tunisians.
Materials and methods
Study subjects
Study subjects comprised 250 unrelated healthy Southern Tunisian individuals of both genders (119 males and 131 females), who were randomly collected and selected from different cities (capitals of governorates) of South Tunisia (Figure 1), taking into account the number of inhabitants in each governorate. All subjects were randomly chosen among individuals whose ancestors have lived in the region for at least three generations. No ethnic, linguistic or religious selection has been applied in the sampling to ensure a representative sample of the current southern population. All participants were interviewed to ensure that no individuals have common ancestry going back at least three generations. Informed and written consent to participate in the study was obtained from all participants; consent being approved by participating institutions. Research and ethics committees of National Blood Transfusion Center (Tunis, Tunisia) and University of Tunis El Manar (Tunis, Tunisia) approved the protocol of the study, as per the Declaration of Helsinki.
DNA extraction
Genomic DNA was prepared from peripheral mononuclear cells using salting-out method.11 Ethylenediaminetetraacetic acid blood samples were lysed and pellets were stored frozen at −20 °C. The cell lysates were digested overnight at 37 °C with 0.2 ml of 10% SDS and 0.5 ml of a proteinase K solution. After digestion was complete, 1 ml of saturated NaCl (~6 m) was added to each tube and shaken vigorously for 15 s, followed by centrifugation at 2500 r.p.m. for 15 min. The supernatant containing the DNA was transferred to another tube. Exactly two volumes of absolute ethanol were added and the tubes inverted several times until the DNA precipitated. Finally, the obtained DNA was allowed to dissolve in distilled water 2 h at 37 °C. Concentration and purity of DNA samples were spectrophotometrically quantified.
HLA DNA genotyping
High-Resolution Genotyping Kits (Innogenetics, ‘fujirebio-Europe’, NV Zwijndrecht, Belgium)12 were used for HLA class I (A, B) and class II (DRB1 and DQB1) genotyping. These kits are based on the reverse dot-blot hybridization principle. Amplified biotinylated DNA material is chemically denatured, and the separated strands are hybridized with specific oligonucleotide probes immobilized as parallel lines on membrane-based strips. Hybridization is carried out in special test troughs, in a water bath and under gentle agitation. After removing any mismatched amplified material by stringent wash, streptavidin conjugated with alkaline phosphatase is added and bound to any biotinylated hybrid formed previously. The incubation of membrane-based strips with a substrate solution containing a BCIP/NBT chromogen leads to the appearance of a purple/brown precipitate. Finally, the reaction is stopped by a wash step, and the reactivity pattern of the probes is recorded and interpreted. In the case of suspected homozygosity or ambiguities, samples will be retyped using One Lambda PCR-SSP High-Resolution Kits (One Lambda, Canoga Park, CA, USA) according to the manufacturer’s protocol. Briefly, PCR reaction was performed by using multiple pairs of cis-located allele-specific primers. After amplification, 10 μl of each PCR reaction was transferred in sequence to a 2.5% agarose gel with 0.5 μg ml−1 ethidium bromide and electrophoresed at 150 V for 4 min. Then, the result of migration is documented by photography. The patterns of positive amplifications were used to interpret HLA genotypes by using an appropriate software. The assignment of HLA alleles was made as per the World Health Organization Nomenclature Committee for Factors of HLA System.13
Statistical analysis
HLA allele frequencies were calculated by the gene counting. Haplotype frequencies were estimated by maximum likelihood from genotypic data using the expectation-maximization algorithm,14 embedded in the Arlequin v.2.0.1 software.15 Linkage disequilibrium (LD) between alleles, defined as the non-random association of two alleles of two loci on the same chromosome, and the level of significance (P) for 2 × 2 comparisons, and the relative LD (D′), were also calculated by Arlequin.16 Phylogenetic trees (dendrograms) were constructed from individual allelic frequencies by the neighbor-joining (NJ) method,17 with standard genetic distances (SGDs),18 using the DISPAN software.19 Three-dimensional correspondence analysis and bi-dimensional representation were carried out using VISTAV5.02 software.20 Correspondence analysis, a geometric technique used for displaying a global view of the relationship among populations according to HLA (or other) allele frequencies, was based on the differential allele frequencies among populations, and on the display of a statistical projection of these differences.
PyPop (Python for Population genomics, version 0.7.0 http://www.pypop.org) was used to perform Hardy–Weinberg testing, pairwise LD estimates21, 22 and Ewens–Watterson homozygosity test.23, 24 This test of homozygosity was applied to each locus, using Slatkin’s Monte Carlo implementation of the exact test. The observed (Fobs) and expected (Fexp) homozygosity (under neutral selection) were calculated, respectively, as sum of the squares of allele frequencies and through simulation, for the same sample size with the same number of alleles. The difference between Fobs and Fexp, divided by the square root of the variance of Fexp provides the normalized deviate of the homozygosity (Fnd).23, 24, 25, 26 The latter was used to infer the action of balancing or directional selection at each locus. The observed homozygosity value for populations evolving under neutral conditions will be similar to the expected homozygosity value, and the resulting Fnd value will be close to zero. Significantly negative Fnd values imply balancing selection and/or high levels of geneflow, whereas significantly positive values imply directional selection and/or extreme demographic effects (e.g., a population bottleneck) as a result of genetic drift.
The correlations between two-locus D′ and physical distance (PD) was assessed using the nonparametric coefficient of Spearman.27 Spearman's rank correlation or Spearman's ρ is a nonparametric test that is used to measure the degree of statistical dependence between two variables. Spearman's rank correlation test does not assume any assumptions about the distribution of the data, and is the appropriate correlation analysis when the variables are measured on a scale that is at least ordinal. Its value ranges from −1 to 1. If ρ is <0, the correlation is negative; if it is >1, the correlation is positive. A perfect Spearman's correlation of +1 or −1 occurs when each of the variables is a perfect monotone function of the other. Spearman's coefficient is appropriate for both continuous and discrete variables, including ordinal variables.
Results
HLA allele frequencies in the studied population
The expected and observed allele frequencies for HLA-A, -B, -DRB1, and -DQB1 loci were in Hardy–Weinberg equilibrium in the population sample (Table 2). The frequencies of HLA-A, -B, -DRB1 and -DQB1 alleles in Southern Tunisians are presented in Table 3. One hundred and thirty-seven different HLA alleles were detected in the sample. Of the thirty-two HLA-A alleles identified, A*02:01 (18.02%), A*34:02 (9.31%) and A*01:01 (8.5%) were the most frequent allele in Southern Tunisians. These were also observed in high frequencies in North African,28 Iberian29 and Mediterranean30 populations. Among HLA-B alleles, 52 were identified in Southerner Tunisians, of which B*50:01 (9.11%) and B*51:01 (7.49%) were the most frequent. Both B*50 and B*51 are common alleles in several Mediterranean and Arab-speaking populations.28, 30, 31, 32, 33, 34, 35, 36, 37
Among HLA class II alleles, 32 DRB1 alleles were found in Southern Tunisians; the most frequent was DRB1*07:01 (22.06%), which was present at high frequencies in Tunisian Berbers (17.6%),33 and from the Ghannouch area (28.7%).10 In addition, DRB1*03:01 (16.4%) was frequent in Tunisians,9, 10, 32, 33, 34, 38 which was also present at comparable frequencies in Moroccans (17.3%),39 Berbers (15.1%)33 and Basques.29 In addition, of the 21 HLA-DQB1 alleles detected, DQB1*02:01 was the most frequent (17.21%), followed by DQB1*02:02 (16.6%) and DQB1*03:01 (16.6%). These alleles were also reported for Tunisian and Mediterranean populations.9, 10, 32, 33, 34
Allelic comparison between Tunisians and other populations
The frequencies of HLA alleles in Southern Tunisians were compared with those of other Arab-speaking, Mediterranean and sub-Saharan populations by high-resolution HLA-DRB1 (Figure 2), generic HLA-B (Figure 3), HLA-DRB1 and -DQB1 (Figure 4) and HLA-A, -B, -DRB1 and -DQB1 data (Figure 5). The latter were performed to confirm our results, as some of the populations included for comparison lacked high-resolution HLA-DRB1 data. This was carried out at the levels of NJ (Figures 2, 3, 4, 5), SGDs (Table 3), and correspondence analysis (Figure 6).
Neighbor-joining dendrogram showing relatedness between Southern Tunisians and other populations. Standard genetic distances (SGDs) between populations were calculated by using high-resolution HLA-DRB1 genotyping. Data from other populations were taken from references detailed in Table 1. Bootstrap values from 1.000 replicates are shown. Only individuals with defined DRB1 subtypes are considered. A full color version of this figure is available at the Journal of Human Genetics journal online.
Neighbor-joining dendrogram showing relatedness between Southern Tunisians and other populations. Standard genetic distances (SGDs) between populations were calculated by using generic HLA-B genotyping. Data from other populations were taken from references detailed in Table 1. Bootstrap values from 1.000 replicates are shown. A full color version of this figure is available at the Journal of Human Genetics journal online.
Neighbor-joining dendrogram showing relatedness between Southern Tunisians and other populations. Standard genetic distances (SGDs) between populations were calculated by using generic HLA-DRB1 and -DQB1 genotyping. Data from other populations were taken from references detailed in Table 1. Bootstrap values from 1.000 replicates are shown. A full color version of this figure is available at the Journal of Human Genetics journal online.
Neighbor-joining dendrogram showing relatedness between Southern Tunisians and other populations. Standard genetic distances (SGDs) between populations were calculated by using generic HLA-A, -B, -DRB1 and -DQB1 genotyping. Data from other populations were taken from references detailed in Table 1. Bootstrap values from 1.000 replicates are shown. A full color version of this figure is available at the Journal of Human Genetics journal online.
Correspondence analysis showing a global view of the relationship among Mediterranean populations according to HLA allele frequencies in three dimensions (bi-dimensional representation). HLA-DRB1 allele frequency data. Only individuals with defined DRB1 subtypes are considered. A full color version of this figure is available at the Journal of Human Genetics journal online.
NJ dendrogram
Results of HLA-DRB1 and -DQB1 (Figure 4) and HLA-A, -B, -DRB1 and -DQB1 data (Figure 5) differ slightly from those obtained by HLA-DRB1 (Figure 2) or HLA-B (Figure 3) data. NJ tree constructed with DRB1 and DQB1, and HLA-A, -B, -DRB1 and -DQB1 allele frequencies shows lower bootstrap values, probably due to lower confidence limits of these NJ trees. NJ dendrograms, using standard SGD based on high-resolution HLA-DRB1 or generic HLA-B data, demonstrated steady gradient of relatedness between Western and Eastern Mediterranean populations. NJ branches, based on high-resolution HLA-DRB1, showed high bootstrap values, and the populations were grouped into two branches. The first was also divided into two sub-branches: one including Southern Tunisians, Spanish populations, North Africans and French, whereas the other included Eastern Mediterraneans (Palestinians, Cretans, Lebanese, Macedonians), Italians and Moroccan Jews. On the other hand, the second branch comprises Greeks and sub-Saharans. Bushmen and Jordanians form an outgroup.
SGDs comparison
SGD based on HLA-DRB allele frequencies indicated that Southern Tunisians are closer to Western than to Eastern Mediterranean populations. This was illustrated in Table 4, in which Gabesians had the closest genetic distance (1.1 × 10−3), followed by Moroccans from Agadir (4.9 × 10−3), Spanish (8.4 × 10−3), Tunisian Berbers (2.15 × 10−2), Libyans (2.80 × 10−2), Algiers (3.11 × 10−2), Basques-Arratia, Moroccans and Northern Tunisians-A. Southern Tunisians appear to be distinct from Eastern Mediterranean populations, including Arab-speaking Palestinians, Jordanians and Lebanese. The same result, with minor difference, was observed using SGD based on HLA-B allele frequencies.
Correspondence analysis
Correspondence analysis, using high-resolution HLA-DRB1 data (Figure 6), shows two main clusters. The first grouped together Western Europeans and North Africans (including Southern Tunisians) and the second combines Eastern Mediterraneans, except for Greeks, who were grouped with sub-Saharan population; Jordanians and Bushmen being outside this grouping scheme. Figure 6 showed that Southern Tunisians are related to North Africans, Iberians and other Western Mediterranean populations.
HLA-A, -B, -DRB1 and -DQB1 LD
HLA haplotype analysis allowed comparison of Southern Tunisians with those previously reported for other populations. Table 5 depicts HLA class I (A, B) and class II (DRB1, DQB1) two-locus haplotypes with significant LD (P<0.05 in all cases) in Southern Tunisians. The most frequent two-locus HLA haplotypes seen in this study were also common in Mediterranean populations (Table 5). Indeed, the DRB1*03:01–DQB1*02:01 haplotype (16.27%), known as an Iberian paleo-North African haplotype, was found in Northern Tunisians (14.08%),32 Tunisian Berbers (11.26%),33 Algerians (11.3%),31 Moroccans (17.3%),28 Chaouya population40, 41 and Basques (17.5%).29 DRB1*07:01–DQB1*02:02, which is the most frequent haplotype in Southern Tunisians (18.02%), was also present in Ghannouchians (16.46%),10 Tunisian Berbers (16.03%),33 Moroccans (12.6%)28, 40, 41 and Spaniards (17.3%).42 Except for the two HLA-DRB1 and -DQB1 haplotypes previously cited, no high frequency of HLA two-locus haplotypes are found in Southern Tunisia. This may be due to the existence of a higher admixture of Mediterraneans in southern population.
HLA class I and class II extended haplotype analysis
Table 6 lists the frequent HLA-A–B–DRB1–DQB1 extended haplotypes detected in Southern Tunisians. The most frequent four-loci haplotype was A*02:01–B*50:01–DRB1*07:01–DQB1*02:02 (3.2%), which was also reported for Northern Tunisians (1.2%),32 Tunisian Berbers (8.1%),33 Gabesian Tunisians (2.6%),9 Moroccan Jews (2%),43 Spaniards (1.2%)40 and Anatolian Turkish (1.3%)44 populations. In addition, A*24:02-B*08:01-DRB1*03:01–DQB1*02:01 haplotype, present in Southern Tunisians (2.33%), is also found in Gabesian Tunisians (1.6%)9 and Ghannouchian Tunisians (4.2%),10 and is frequently associated with A*01 (instead of HLA-24) in many Mediterranean populations, including Spaniards (3.4%) and Basques (5%).42, 45, 46 Other HLA-A–B–DRB1–DQB1 extended haplotypes found in the southern population indicate a Mediterranean background.
Global LD estimates
Table 7 shows global LD estimates of the associations between HLA loci. All pairs of loci show significant LD. DRB1:DQB1 has the strongest association (D′=0.90507). These genes lie adjacent to one another in the major histocompatibility complex separated by a reduced PD of ~0.062 Mb. On the contrary, the A:DQB1 LD estimates have the lowest values (D′=0.54772). This is because of big PD (~2.65 Mb) separating HLA-A and HLA-DQB1 genes, which promotes the increase of recombination rate. However, A:B shows slightly higher LD value (D′=0.67995; PD=1.4 Mb) compared with B:DQB1 (D′=0.67865; PD=1.24 Mb). This was observed in the 13th Workshop Anthropology project and others works,47, 48 and may be result from the low levels of polymorphism seen at HLA-DQB1, relative to the HLA-A and -B loci. A significant negative correlation, using the nonparametric coefficient of Spearman, was found between two-locus D′ and PD (r=−0.94286; P=0.0048). This result indicates that the variation of the LD strength is inversely proportional to the PD separating two loci (Figure 7). This is owing to the fact that the rate of recombination increases with the PD.
Ewens–Watterson homozygosity test of neutrality
The results of the Ewens–Watterson homozygosity test are shown in Table 8. No significant deviation was found for any of the genes analyzed, although homozygosity was usually lower than expected (negative Fnd values) under selective neutrality. In addition, the significant differences between observed and expected homozygotes for each locus (except for HLA-A) indicates an overall significant trend away from the null hypothesis of neutral evolution (HLA-A, P-value: 0.1654; HLA-B, P-value: 0.0287; HLA-DRB1, P-value: 0.0233; HLA-DQB1, P-value: 0.0073), suggesting that the allele frequency distributions at all four loci have been shaped by balancing selection. This trend is frequently observed for the classical HLA genes, being an evidence of balancing selection. It is worth pointing out that very large population samples are needed to obtain statistical significance if selection pressure is low, as estimated for the HLA genes.49
Discussion
To the best of our knowledge, this was the first anthropological study that investigated HLA genetic profiles (high resolution) of Southern Tunisians. Indeed, there are several major differences characterizing our study compared with previous works carried out in Tunisia. First, all individuals of this present study are molecularly typed with high-resolution kits for all studied HLA markers. The earlier studies have used a generic low-resolution typing for HLA class I genes,10, 32 and even sometimes samples were serologically typed, especially for HLA class I genes.48 Second, our study has recruited a large number of sample (250 individuals) belonging to different governorates of the South what makes it more representative of the total current southern population. However, previous studies have investigated some isolates existing in the South,10, 33, 50, 51 and their number of samples does not exceed one hundred. Finally, a major of the HLA studies carried out in all Tunisia are closer to a simple presentation of the distributions of HLA alleles in studied populations,38, 52, 53 while this present work uses means of anthropological and evolutionary analysis (dendrograms, correspondence analysis, genetic distance, study of selection and so on). All these characteristics make our study a unique anthropological study in the South.
The testing for Hardy–Weinberg equilibrium based on the high-resolution typing data of the entire population showed that individual HLA-A, -B, -DQB1 and -DRB1 phenotypes were in Hardy–Weinberg equilibrium. Besides, the frequencies of HLA-A and -B haplotypes and D' exhibit low values in Southern Tunisians. A similar observation has already been reported for the North Africans and the majority of the studied populations (with the exception of some isolates), especially when compared with those of HLA-DRB1 and -DQB1 haplotypes. This observation is mainly explained by the relatively high PD between A and B loci, which increases genetic recombination and subsequently diversity. All pairs of loci show significant LD, and the presence of significant correlation between two-locus D′ and PDs. The four loci have negative Fnd values indicating an overall direction toward balancing selection, that is, selection for sustaining high diversity of the HLA alleles in this population. A large sample is needed to obtain statistical significance if selection pressure is low. It has been previously shown54 that there is evidence of balancing selection in HLA loci (A, C, B, DRB1, DQA1 and DQB1) from 497 human populations across the globe. DQA1 displays the strongest evidence for balancing selection that was attributed to selection for variability in the amino-acid residues that form antigen-binding/presenting pocket. Only DPA1 and DPB1 demonstrate positive Fnd values in some regions, which may be due to the low power of these statistical tests at loci with few population samples. There are evidences that the evolution of the HLA polymorphism is complex, with multiple stochastic and deterministic evolutionary factors acting in concert,55 so that the demonstration of each of these factors still is a difficult task.
HLA class I and class II genotype distribution among Southern Tunisians were compared with those of Mediterranean, Arab-speaking and sub-Saharan African communities using genetic distances, NJ dendrograms, correspondence and haplotype analysis. We included HLA class II genotypes in the comparison between populations (NJ trees, correspondence analysis and SGD), as the informativeness of HLA class II loci is much higher compared with those of class I.32, 56 It should be noted that several populations, especially neighboring, were not typed for HLA class II loci, and only generic (sometimes only serological data) are available, which reduced the number of populations used in later comparisons.
Southern Tunisians, North Africans and Iberians
Table 4 shows that Gabesians are the closest Tunisians to Southerners (regardless of 'Tunisian' population, because it represents all Tunisia). This is compatible with the geography, as Gabes region is a part of the South. On the contrary, the genetic distance (SGD), using HLA-DRB1, between Southern Tunisians and Ghannouchians (belonging to Eastern South of Tunisia) is higher compared with other populations (Libyans, Algiers and on on). This observation can be explained by the high frequency of HLA-DRB1*0701 (28.6%) in the population of Ghannouch. Indeed, this frequency is one of the highest observed among all populations tested.10 In addition, this small village is characterized by high endogamy, and behaves as an isolate.
Our results show that Southern Tunisians are related to North Africans, which is attributed to the sharing of similar history by North Africans, albeit with minor differences. Indeed, native Berbers were successively invaded by populations from predominantly Mediterranean communities. Later, admixture of North Africans (including Tunisians) was brought about by the Muslim conquest of North Africa (7th century ad), and the massive Bedouin immigration (11th century), followed by Spanish (16th century) and French (19th–20th century).57, 58 Correspondence analysis, NJ trees, SGD and haplotype studies showed that Southern Tunisians are related to Basques and Spaniards. Several historic events can support the relatedness between North Africans and Iberians. First, this relatedness can be attributed mainly to the northward Saharan migration, which likely occurred in 10 000–4000 bc, when the Berbers relocated to the Northern Mediterranean coast during hyperarid conditions.59 Second, it can be also explained by the similar history between Iberians and North Africans, as both were invaded by Phoenicians, Romans, Germans (Visigoths in Iberia, Vandals in North Africa), Muslim Arabs and Berbers.60 In this similar history, there was an important geneflow. Indeed, during the Muslim invasion of Iberia in the 8th century ad, this invasion was launched from North Africa and the majority of the recruited invaders were Berbers. North African Berber Muslims settled almost eight centuries on Spain, and modern studies estimate more than a million of Moriscos have integrated into the Iberian society. However, the Moriscos were subject to systematic expulsions from Spain's various kingdoms between 1609 and 1627ad. Several studies, being based on the number of recorded expulsion edicts, have shown that the total number of deportees was estimated at 300 000 Moriscos.61
We think that this systematic expulsion could not alter the relatedness between the Iberian and North Africa for several reasons: first, this relatedness was mainly due to prior factors to Islam invasion (the sub-Saharan migration described above). Second, it is difficult even impossible to eradicate an ethnic group and its genetic traces after eight centuries of settlement and integration. Third, the large majority of those permanently expelled settled on the western fringe of the Ottoman Empire and the Kingdom of Morocco (North Africa), which homogenized both populations and increased their relatedness. Finally, the expulsion was made on a religious basis and so there were so many Muslims of Iberian origin.
Several studies using single-nucleotide polymorphism data show that human genetic diversity in southern Europe is higher compared with that in other regions of the continent. This difference has been partially attributed to gene flow from Africa. In addition, most disease risk alleles from genome-wide association studies follow expected patterns of divergence between Europe and North Africa.62 Other study shows that gene flow across the Strait of Gibraltar occurred at relatively high rates since pre-Neolithic times.63 These observations support the relatedness between Iberians and North Africans.
Southern Tunisians, Berbers and Eastern Arabs
Our NJ trees, correspondence analysis, SGD and haplotype studies support the relatedness of Southern Tunisians to Berbers. Indeed, these analyses were consistent with geography and ancestry. This relatedness can be explained by native aspect of the Berbers, and their present concentration in Southern Tunisian governorates and mountains. This was suggested to be the result of the migration of Berbers to Southern Tunisia mountainous regions, where they took refuge from invaders, in particular Arabs.8 In addition, Tunisians are distinct from Palestinians, Lebanese and Jordanians. This is probably because of the influx from the Middle East was low compared with established Berbers. This low Arab contribution into Tunisian genetic pool is also explained by the low admixture between Berbers and Arab tribes, as most Berbers resided in the mountains from the fear of persecution. Cultural barriers, such as language, religion, traditions, between Berbers and Arabs were added to the low admixture. The other major reason explaining the low contribution of Arabs to the southern genetic pool was that the most part of the Banu Hilal and Banu Soulaym invaders were of Berber origin. Indeed, a large number of Egyptian Berbers have been recruited and Arabized by these tribes in Upper Egypt (region of ancient Egypt in the valley of the Nile River south of the delta area) where they were installed for a long time. In addition, several references reported that the number of individuals of these tribes has doubled many times after their arrival in the Upper Egypt.8 It is worth noting that about two hundred and fifty thousand Arabs (estimated at fifty thousand the number of warriors and two hundred thousand the number of Bedouin) belonging to the Banu Hilal and Banu Sulaym, Arabian Bedouin tribes originating from the Hijaz and Nejd, migrated in several massive waves from Upper Egypt into Tunisia. However, some references indicated that the number of members of these tribes, when they arrived Upper Egypt, did not exceed few thousands.64 Consequently, this invasion could not deeply modify the genetic pool of Southern population. Therefore, Berber genetic profile of Southern Tunisians remains evident, although the main focus of Arab invasion of North Africa was Southern Tunisia, which constituted the gateway for Arab tribes invading North Africa cities.
This study put Jordanians out of tested groups. It was expected that the Jordanians associate with the Palestinians in the western Mediterranean cluster because both are neighbors sharing almost the same history, and besides, a large number of Palestinian refugees live in Jordan since 1948.65 Indeed, some unofficial censuses estimate that Palestinians constitute more than half of the Jordanian population. In addition, the Palestinian West Bank was occupied by Jordan from 1951 until the Six-Day War (1967).65 Today, most Palestinians and their descendants in Jordan are fully naturalized, making Jordan the only Arab country to fully integrate the Palestinian refugees of 1948. All these reasons indicate the need for further studies to confirm this result.
It is interesting to note that the association of Jordanians and Bushmen (Figure 2) with western populations is due to a mismatch (which is clearer in NJ trees, data not shown). Indeed, the dendrograms have given under UPGMA form (Unweighted Pair Group Method with Arithmetic Mean). The latter is the simplest method for constructing trees, and easier to read and analyze. It presents appropriately the different clusters. However, mismatch and false associations are difficult to detect, and it is very sensitive to unequal evolutionary rates; not reliable if data are not ultrametric. This is because UPGMA assumes the same evolutionary speed on all lineages. This would mean that all leaves (terminal nodes) have the same distance from the root. In reality, the individual branches are very unlikely to have the same length. Therefore, UPGMA frequently generates inaccuracies in tree topologies.
Southern Tunisians, Blacks and sub-Saharans
Tunisian Blacks derive their origin from a large area stretching from West Africa to Lake of Chad. The kingdoms of Bornu (North of Chad), Fezzan and Ghadames regions (South West of Libya) provided the majority of Black caravans to Southern Tunisia.6 Trans-Saharan trade provided the bulk of the African Blacks flow. A secondary Black flow came from the Arab invasions of the region or from Europe during the period of the colonization of the Maghreb (North-West Africa). While their official number remains unknown, Tunisian Blacks are more frequent in Southern than in Northern Tunisia, and exist in large numbers in most Southern governorates, such as Gabes, Kebili, Tozeur and Tataouine. However, HLA data showed that Southern Tunisians are related to Northern Tunisians, with a big genetic distance from sub-Saharan populations. This suggests that the Black contribution to Southern genetic pool is little, probably because of the high endogamy in Black populations, as interethnic marriages are rare because of social barriers.
This study shows that sub-Saharans are distinct from Tunisians and other Mediterranean populations, and tend to cluster only with Greeks (Figures 2 and 6).10, 56, 66 In addition, several specific Greek alleles were detected in some West African (Rimaibe, Fulani and Mossi) and East African (Oromo, Amhara and Nubians) tribes. Besides, Greeks are the only Caucasoid population who bears cystic fibrosis mutations typical of Black Africans.67 This suggests an admixture between the Greeks and sub-Saharans at an ancient time, and it was suggested that this admixture has occurred during Egyptian pharaonic times.64, 66, 68 However, other studies using the HLA-DRB1 marker did not detect this relationship.69 It may be due to that the author did not use, in the comparison, the same populations of sub-Saharan origin. It should also be noted that this probable relationship between Greeks and sub-Saharans is not so far demonstrated by anthropological studies using HLA class I (Figure 3) or non-HLA markers.
Finally, our results using HLA genes depict that all Tunisian populations (Berber, Southern and Northern populations) are closely related, and show a clear relatedness to North Africans, Iberians and Western Mediterraneans, but they are distinct from sub-Saharans and Eastern Arabs. These results are in agreement with those previously carried out in the region.9, 10, 31, 32, 33, 34, 37, 59, 66
It is important to note that some Tunisian studies, using Y-chromosome single-nucleotide polymorphisms,70 and mitochondrial markers71, 72 in various Tunisian ethnic groups showed that Tunisian populations were characterized by a highly genetic heterogeneity because of isolation and genetic drift. In these studies, where the relatedness among Tunisian populations is not obvious, there were disparities and even sometimes a discrepancy between paternal and maternal lineage results concerning the degree of relatedness to sub-Saharans, Eurasians and eastern Arabs. Indeed, some studies using paternal lineage (Y-chromosome markers) revealed that no major sub-Saharan African or European influence was found, which contrasts with previous studies showing a high amount of sub-Saharan and Eurasian maternal lineages (mitochondrial markers).70 On the contrary, the results using HLA markers are more homogeneous, which may indicate that HLA genes, and especially HLA-DRB1, correlate better with geography, which proves its precious informativeness.
In conclusion, even with the presence of cultural and ethnic differences, Southern Tunisians show a close relatedness to others Tunisian populations, including Berbers, North Africans and Iberians. Surprisingly, Southern Tunisians are distinct from other Middle Eastern Arabs (Palestinians, Lebanese and Jordanians) and sub-Saharans, despite the Arab successive incursions, the strong Arabization that occurred in Southern Tunisia, and the 18th–19th century Black caravans. This result was confirmed by all analyses carried out in this study (genetic NJ trees, correspondence analysis, HLA genetic distances and haplotypes).
References
Shiina, T., Hosomichi, K., Inoko, H. & Kulski, J. K. The HLA genomic loci map: expression, interaction, diversity, and disease. J. Hum. Genet. 54, 15–39 (2009).
Robinson, J., Soormally, A. R., Hayhurst, J. D. & Marsh, S. G. The IPD-IMGT/HLA database—new developments in reporting HLA variation. Hum. Immunol. 77, 233–237 (2016).
Vina, M. A., Hollenbach, J. A., Lyke, K. E., Sztein, M. B., Maiers, M., Klitz, W. et al. Tracking human migrations by the analysis of the distribution of HLA alleles, lineages and haplotypes in closed and open populations. Philos. Trans. R. Soc. Lond. Ser. B 367, 820–829 (2012).
Brett, M. & Fentress, E. The Berbers, (Blackwell Publishers, Oxford, UK, 1997).
Stearns, P. N. & Leonard, W. L. in The Encyclopedia of World History: Ancient, Medieval, and Modern, Chronologically Arranged 6th edn, 129–131 (Houghton Mifflin Harcourt, New York, NY, USA, 2001).
Austin, R. A. The Transaharian Slave Trade. Essays in the Economic History of the Atlantic Slave Trade, (New York Academy Press, New York, NY, 1979).
Lucette, V. & Abraham, L. U. Juifs en terre d’islam: les communautés de Djerba 13 (éd. Archives contemporaines, Paris, 1991).
Ibn Khaldūn, A. in The Muqaddimah: An Introduction to History (Trans. Franz Rosenthal, ed. Dawood, N. J., 1967) (abridged).
Hajjej, A., Hajjej, G., Almawi, W. Y., Kaabi, H., El-Gaaied, A. & Hmida, S. HLA class I and class II polymorphism in a population from south-eastern Tunisia (Gabes Area). Int. J. Immunogenet. 38, 191–199 (2011).
Hajjej, A., Hmida, S., Kaabi, H., Dridi, A., Jridi, A., El Gaaled, A. et al. HLA genes in Southern Tunisians (Ghannouch area) and their relationship with other Mediterraneans. Eur. J. Med. Genet. 49, 43–56 (2006).
Miller, S. A., Dykes, D. D. & Polesky, H. F. A sample salting out procedure for extraction DNA from human nucleated cells. Nucleic Acids Res. 16, 1215–1218 (1988).
Buyse, I., Decorte, R., Baens, M., Cuppens, H., Semana, G., Emonds, M. P. et al. Rapid DNA typing of class II HLA antigens using the polymerase chain reaction and reverse dot blot hybridization. Tissue Antigens 41, 1–14 (1993).
Marsh, S. G. E., Albert, E. D., Bodmer, W. F., Bontrop, R. E., Dupont, B., Erlich, H. A et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75, 291–455 (2010).
Excoffier, L. & Slatkin, M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 12, 921–927 (1995).
Schneider, S., Kueffer, J. M., Roessli, D. & Excoffier, L. Arlequin: A Software Environment for the Analysis of Population Genetics Data, (Genetics and Biometry Lab, Geneva, Switzerland, 1996).
Imanishi, T., Akaza, T., Kimura, A., Tokunaga, K., Gojrobi, T. in HLA 1991 Vol. I (eds Tsuji, K., Aizawa, M. & Sasazuki, T.) 76–79 (Oxford University Press, Oxford, UK, 1992).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Nei, M. Genetic distances between populations. Am. Nat. 106, 283 (1972).
Nei, M., Tajima, Y. & Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J. Mol. Evol. 19, 153–170 (1983).
Young, F. W. & Bann, C. M. in Statistical Computing Environments for Social Researches (eds Stine, R. A. & Fox, J.) 207–236 (Sage Publications, New York, NY, 1996).
Lancaster, A. K., Nelson, M. P., Single, R. M., Meyer, D. & Thomson, G. in Pacific Symposium on Biocomputing Vol. 8 (eds Altman, R. B., Dunker, K., Hunter, L., Jung, T. & Klein, T.) 514–525 (World Scientific, Singapore, Singapore, 2003).
Lancaster, A. K., Single, R. M., Solberg, O. D., Nelson, M. P. & Thomson, G. PyPop update—a software pipeline for large-scale multilocus population genomics. Tissue Antigens 69, 192–197 (2007).
Ewens, W. The sampling theory of selectively neutral alleles. Theor. Pop. Biol. 3, 87–112 (1972).
Watterson, G. The homozygosity test of neutrality. Genetics 88, 405–417 (1978).
Slatkin, M. An exact test for neutrality based on the Ewens sampling distribution. Genet. Res. 64, 71–74 (1994).
Slatkin, M. A correction to the exact test based on the Ewens sampling distribution. Genet. Res. 68, 259–260 (1996).
Glantz, S. A. Primer of Biostatistics 7th edn (McGraw-Hill, New York, NY, 2012).
Gomez-Casado, E., del Moral, P., Martinez-Laso, J., García-Gómez, A., Allende, L., Silvera-Redondo et al. HLA gene in Arabic-Speaking Moroccans: close relatedness to Berbers and Iberians. Tissue Antigens 55, 239–249 (2000).
Comas, D., Mateu, E., Calafell, F., Pérez-Lezaun, A., Bosch, E., Martínez-Arias, R. et al. HLA class I and class II DNA typing and the origin of Basques. Tissue Antigens 51, 30–40 (1998).
Clayton, J. & Lonjou, C. in Genetic Diversity of HLA. Functional and Medical Implications Vol. 1 (ed. Charron, D.) 665–820 (EDK: Paris, 1997).
Arnaiz-Villena, A., Benmamar, D., Alvarez, M., Diaz-Campos, N., Varela, P., Gomez-Casado, E. et al. HLA allele and haplotype frequencies in Algerians. Relatedness to Spaniards and Basques. Hum. Immunol. 43, 259–268 (1995).
Hajjej, A., Kâabi, H., Sellami, M. H., Dridi, A., Jeridi, A., El Borgi, W. et al. The contribution of HLA class I and II alleles and haplotypes to the investigation of the evolutionary history of Tunisians. Tissue Antigens 68, 153–162 (2006).
Hajjej, A., Sellami, M. H., Kaabi, Hajjej, G., El-Gaaied, A., Boukef, K. et al. HLA class I and class II polymorphisms in Tunisian Berbers. Ann. Hum. Biol. 38, 156–164 (2011).
Hajjej, A., Almawi, W. Y., Hattab, L., El-Gaaied, A. & Hmida, S. HLA class I and class II alleles and haplotypes confirm the Berber Origin of the Present Day Tunisian Population. PLoS ONE 10, e0136909 (2015).
Sánchez-Velasco, P., Karadsheh, N. S., García-Martín, A., Ruíz de Alegría, C. & Leyva-Cobián, F. Molecular analysis of HLA allelic frequencies and haplotypes in Jordanians and comparison with other related populations. Hum. Immunol. 62, 901–909 (2001).
Galgani, A., Mancino, G., Martínez-Labarga, C., Cicconi, R., Mattei, M., Amicosante, M. et al. HLA-A, -B and -DRB1 allele frequencies in Cyrenaica population (Libya) and genetic relationships with other populations. Hum. Immunol. 74, 52–59 (2013).
Arnaiz-Villena, A., Elaiwa, N., Silvera, C., Rostom, A., Moscoso, J., Gómez-Casado, E. et al The origin of Palestinians and their genetic relatedness with other Mediterranean populations [retraction] Hum. Immunol. 62, 889–900 (2001).
Hmida, S., Gauthier, Dridi, A., Quillivic, F., Genetet, B., Boukef et al. HLA class II gene polymorphism in Tunisians. Tissue Antigens 45, 63–68 (1995).
Izaabel, H., Garchon, H. J., Caillat-Zucman, S., Beaurain, G., Akhayat, O., Bach, J. F. et al. HLA class II DNA polymorphism in a Moroccan population from the Souss, Agadir area. Tissue Antigens 51, 106–110 (1998).
Canossi, A., Piancatelli, D., Aureli, A., Oumhani, K., Ozzella, G., Del Beato, T. et al. Correlation between genetic HLA class I and II polymorphisms and anthropological aspects in the Chaouya population from Morocco (Arabic speaking). Tissue Antigens 76, 177–193 (2010).
Brick, C., Atouf, O., Bouayad, A . & Essakalli, M. Moroccan study of HLA (-A, -B, -C, -DR, -DQ) polymorphism in 647 unrelated controls: Updating data. Mol. Cell. Probes 29, 197–207 (2015).
Martinez-Laso, J., De Juan, Martinez-Quiles, N., Gomez-Casado, E., Cuadrado, E. & Arnaiz-Villena, A. The contribution of the HLA-A, -B, -C and -DR, -DQ DNA typing to the study of the origins of Spaniards and Basques. Tissue Antigens 45, 237–245 (1995).
Roitberg-Tambur, A., Witt, C. S., Friedmann, Safirman, C., Sherman, Battat, S. et al. Comparative analysis of HLA polymorphism at the serologic and molecular level in Moroccan and Ashkenazi Jews. Tissue Antigens 46, 104–110 (1995).
Arnaiz-Villena, A., Karin, M., Bendikuze, N., Gomez-Casado, E., Moscoso, J., Silvera, C. et al. HLA alleles and haplotypes in the Turkish population: relatedness to Kurds, Armenians and other Mediterraneans. Tissue Antigens 57, 308–317 (2001).
Imanishi, T., Akaza, T., Kimura, A., Tokunaga, K. & Gjobori, T. in HLA 1991 Vol. 1 (eds Tsuji, K., Aizawa, M. & Sasazuki, T.) 1065–1220 (Oxford University Press, Oxford, UK, 1992).
Sanchez-Velasco, P., Gomez-Casado, E., Martinez-Laso, J., Moscoso, J., Zamora, J., Lowy, E. et al. HLA alleles in isolated populations from North Spain: origin of the Basques and the ancient Iberians. Tissue Antigens 61, 384–392 (2003).
Single, R., Meyer, D., Mack, S. J., Lancaster, A., Nelson, M. P. & Erlich, H. et al. in Immunobiology of the Human MHC (ed. Jansen, J. A.) 705–746 (International Histocompatibility Working Group Press, Seattle, WA, 2006).
Mack, S. J., Tu, B., Lazaro, A., Yang, R., Lancaster, A. K., Cao, K. et al. HLA-A, -B, -C, and -DRB1 allele and haplotype frequencies distinguish Eastern European Americans from the general European American population. Tissue Antigens 73, 17–32 (2009).
Satta, Y., O’hUigin, C., Takahata, N. & Klein, J. Intensity of natural selection at the major histocompatibility complex loci. Proc. Natl Acad. Sci. USA 91, 7184–7188 (1994).
Abdennaji Guenounou, B., Yacoubi Loueslati, B., Buhler, S., Hmida, S., Ennafaa, H., Khodjet-Elkhil, H. et al. HLA class II genetic diversity in Southern Tunisia and the Mediterranean area. Int. J. Immunogenet. 33, 93–103 (2006).
Fadhlaoui-Zid, K., Buhler, S., Dridi, A., Benammar El Gaaied, A. & Sanchez-Mazas, A. Polymorphism of HLA class II genes in Berbers from Southern Tunisia. Tissue Antigens 76, 416–420 (2010).
Mahfoudh, N., Ayadi, I., Kamoun, A., Ammar, R., Mallek, B. & Maalej, L. et al. Analysis of HLA-A, -B, -C, -DR, -DQ polymorphisms in the South Tunisian population and a comparison with other populations. Ann Hum Biol. 40, 41–47 (2013).
Ayed, K., Ayed-Jendoubi, S., Sfar, I., Labonne, M. P. & Gebuhrer, L. HLA class-I and HLA class-II phenotypic, gene and haplotypic frequencies in Tunisians by using molecular typing data. Tissue Antigens 64, 520–532 (2004).
Solberg, O. D., Mack, S. J., Lancaster, A. K., Single, R. M., Tsai, Y. & Sanchez-Mazas, A. et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 69, 43–464 (2008).
Apanius, V., Penn, O. & Slev, P. R. et al. The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 15, 179–224 (1997).
Arnaiz-Villena, A., Gomez-Casado, E. & Martinez-Laso, J. Population genetic relationships between Mediterranean populations determined by HLA allele distribution and a historic perspective. Tissue Antigens 60, 111–121 (2002).
Julien, C. A. Histoire de l’Afrique du Nord (Masson et Cie, Paris, 1953).
Murdok, G. P. Africa, Its Peoples and Their Cultural History (McGrawHill, New York, NY, 1959).
Arnaiz-Villena, A., Iliakis, P., González-Hevilla, M., Longás, J., Gómez-Casado, E., Sfyridaki, K. et al. The origin of Cretan populations as determined by characterization of HLA alleles. Tissue Antigens 53, 213–226 (1999).
Fischer, T. in The International Geography (ed. Mill, H. R.) 368–377 (Appleton and Company, New York, NY and London, UK, 1920).
Stallaert, C. Ethnogenesis and Ethnicity in Spain: A Historical–Anthropological Approach to Casticismo (Proyecto A: Barcelona, Spain, 1998).
Botigué, L. M., Henn, B. M., Gravel, S., Maples, B. K., Gignoux, C. R. & Corona, E. et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc. Natl Acad. Sci. USA 110, 11791–11796 (2013).
Currat, M., Poloni, E. S. & Sanchez-Mazas, A. Human genetic differentiation across the Strait of Gibraltar. Evol. Biol. 10, 237 (2010).
Le Bon, G. Arab Civilisation (Libairie Firmin Diderot, France, 1884).
Mark, A. & Tessler, A. History of the Israeli-Palestinian Conflict Vol. 329 (Indiana University Press, Bloomington, IN, 1994).
Arnaiz-Villena, A., Dimitroski, K., Pacho, A., Moscoso, J., Gómez-Casado, E. & Silvera-Redondo, C. et al. HLA genes in Macedonians and the Sub-Saharan origin of the Greeks. Tissue Antigens 57, 118–127 (2001).
Dork, T., El-Harith, E. -H. A., Stuhrmann, M., Macek, M. Jr, Egan, M., Cutting, G. R. et al. Evidence for a common ethnic origin of cystic fibrosis mutation 3120+1G-to-A in diverse populations. Am. J. Hum. Genet. 63, 656–662 (1998).
Herodotus. History (Gredos, Madrid, Spain, 1989).
Petlichkovski, A., Efinska-Mladenovska, O., Trajkov, D., Arsov, T., Strezova, A. & Spiroski, M. High-resolution typing of HLA-DRB1 locus in the Macedonian population. Tissue Antigens 64, 486–491 (2004).
Fadhlaoui-Zid, K., Martinez-Cruz, B., Khodjet-el-khil, H., Mendizabal, I., Benammar-Elgaaied, A. & Comas, D. Genetic structure of Tunisian ethnic groups revealed by paternal lineages. Am. J. Phys. Anthropol. 146, 271–280 (2011).
Fadhlaoui-Zid, K., Plaza, S., Calafell, F., Ben Amor, M., Comas, D. & Bennamar El gaaied, A. Mitochondrial DNA heterogeneity in Tunisian Berbers. Ann. Hum. Genet. 68, 222–233 (2004).
Frigi, S., Cherni, L., Fadhlaoui-Zid, K. & Benammar-Elgaaied, A. Ancient local evolution of African mtDNA haplogroups in Tunisian Berber populations. Hum. Biol. 82, 367–384 (2010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hajjej, A., Almawi, W., Hattab, L. et al. The investigation of the origin of Southern Tunisians using HLA genes. J Hum Genet 62, 419–429 (2017). https://doi.org/10.1038/jhg.2016.146
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.146
This article is cited by
-
Contribution of HLA class I (A, B, C) and HLA class II (DRB1, DQA1, DQB1) alleles and haplotypes in exploring ethnic origin of central Tunisians
BMC Medical Genomics (2024)
-
HLA-A, -B, -C, -DRB1 and -DQB1 allele and haplotype frequencies in Lebanese and their relatedness to neighboring and distant populations
BMC Genomics (2022)