Abstract
Apolipoprotein E (APOE), translocase of outer mitochondrial membrane 40 homolog (TOMM40) and apolipoprotein C-I (APOC1) may extend lifespan by marked delay or escape from age-related diseases. This study aimed to elucidate the association of human longevity with genetic variations in TOMM40/APOE/APOC1 region in a Chinese population. Ten tag single-nucleotide polymorphisms (SNPs) in the TOMM40/APOE/APOC1 region were successfully genotyped in 616 unrelated long-lived individuals and 846 younger controls. Of the 10 SNPs, rs7254892 in 5′ upstream of TOMM40 showed significant association with human longevity (G/A–A/A vs G/G: odds ratio (OR)=1.59, 95% confidence interval (CI)=1.20–2.09, P=0.0011, Bonferroni corrected P (Pc)=0.033). The haplotype analysis suggested that individuals carrying the haplotype A–A–A–A–T–A–T–G–C–A (rs7254892–rs157580–rs2075649–rs2075650–rs157582–rs8106922–rs1160985–rs405697–rs439401–rs445925) tended to have longer lifespan than those carrying the most common haplotype G–G–A–A–C–A–C–A–T–G (OR=1.59, 95% CI=1.19–2.12, P=0.0018, Pc=0.0216). These findings indicated that variants in TOMM40/APOE/APOC1 region might be associated with human longevity. Further studies are needed to identify the causal genetic variants influencing human longevity.
Similar content being viewed by others
Introduction
Human longevity is a complex and multifactorial trait that is influenced by environmental and genetic factors.1 Centenarians are characterized by marked delay or escape from age-related diseases, such as coronary artery disease (CAD), cerebrovascular disease (CVD) and Alzheimer’s disease (AD), which are the leading causes of death. Thus one can suggest that environmental and genetic factors likely to be involved in these disorders may have a role in human longevity.2
The genetic component of human lifespan variation was estimated at ~25% in twin studies.3, 4 Apolipoprotein E (APOE), which encodes the protein apolipoprotein E, is the first to be successfully replicated as one important in exceptional longevity.5 It is one of the most convincing human longevity loci. Variants in APOE have frequently, but not always, been proved to be associated with longevity.2, 6, 7 Although the mechanism is not entirely clear, this association could be explained by marked delay or escape from age-related diseases, such as vascular disease and Alzheimer’s disease, via the effects of APOE on (1) amyloid-β (Aβ) metabolism, (2) neurons or glial cells, including neuronal survival and neurite extension or (3) lipoprotein metabolism.2, 8
Furthermore, genome-wide association studies for human longevity have identified genome-wide significant loci in the translocase of outer mitochondrial membrane 40 homolog (TOMM40) gene,9 and in 3′ downstream of the apolipoprotein C-I (APOC1) gene.10, 11 TOMM40 is adjacent and ~2 kb upstream to APOE, and APOC1 is adjacent and ~5 kb downstream to APOE.
TOMM40 encodes TOM40 protein, a subunit of the multisubunit translocase of the outer mitochondrial membrane, which has a role in the transport of cytoplasmic peptides and proteins into mitochondria. The TOMM40 gene polymorphisms has been widely examined in different AD association studies although the results are contradictory.12, 13 The amyloid precursor protein (APP) forms stable complexes with the TOM40 import channel or with both mitochondrial TOM40 and the translocase of the inner mitochondrial membrane 23 (TIM23) import channel TIM23, and accumulates exclusively in human AD brains but not in age-matched controls, which are the mechanisms for mitochondrial dysfunction by APP in AD.14 Furthermore, the Aβ peptide, a peptide considered to be of major significance in AD, was reported to be transported into mitochondria via the TOM complex and accumulated in the mitochondrial cristae.15 Collectively, there is both genetic and physiological evidence for TOMM40 in AD risk or pathogenesis, which suggests TOMM40 may be also implicated in the process of human longevity. However, until now, few studies of human longevity have been performed for TOMM40 especially in Asians.
APOC1, encoded by APOC1, is a member of apolipoprotein family. In mice, human APOC1 overexpression impairs cognitive functions independent of APOE expression.16 The absence of APOC1 expression also leads to impaired memory functions.17 Thus APOC1 may have a modulatory role in the development of AD. APOC1, together with APOE, take part in a wide variety of biological processes of cholesterol metabolism, membrane remodeling, neuronal apoptosis and reorganization.18, 19 A number of studies have shown that APOC1 is also an important risk factor for AD and observations associating risk factors for cardiovascular disease such as high low-density lipoprotein (LDL) cholesterol, low high-density lipoprotein (HDL) cholesterol and high triglycerides.20, 21, 22, 23 Thus, APOC1 variants might also be involved in human longevity. However, so far, only three studies have reported APOC1 with human longevity and were conducted only in Caucasians.10, 11, 24
In this paper, we hypothesized that common variants in TOMM40 and APOC1, in addition to APOE, may contribute to longevity and conducted an association analysis of TOMM40/APOE/APOC1 locus with human longevity in long-lived cases (⩾98 years) and younger controls (30–70 years) in a Chinese population.
Materials and methods
Study population
The study sample comprised 616 unrelated Chinese long-lived individuals (LLIs) and 846 younger controls. All LLIs were ⩾98 years of age at the time of enrollment (mean age: 102.4±2.3 years, 38 Li and 578 Han people, 102 males and 514 females). The gender ratio in the LLIs was 83.4 females vs 16.6 males, and 93.8% of the LLIs were Han Chinese. The 846 Chinese control subjects were between 30 and 70 years of age (mean age: 48.9±10.6 years, 69 Li and 777 Han people, 159 males and 687 females) and matched the LLIs by gender, ethnical ancestry and geographical origin. All subjects were recruited from Hainan Island between 2009 and 2014. According to the sixth Chinese census database in 2010, among China’s 31 provinces, autonomous regions and municipalities, the highest number of centenarians per 10 000 inhabitants aged 65 years or older is in Hainan (16.64), followed by Guangxi (7.00), Guangdong (6.07), Xinjiang (5.25), Shanghai (3.98).25 All subjects gave informed, written consent prior to participation. The study was approved by the Ethics Committee of Hainan Medical College and by the local data protection authorities.
SNP selection and genotyping
Eleven tag single-nucleotide polymorphisms (SNPs), which span the TOMM40/APOE/APOC1 region (chromosome 19: 44886237-44919549 33.31 kbp, human genome reference assembly GRCh38/hg38), were selected from the phase III Han Chinese in Beijing (CHB) and Southern Han Chinese (CHS) populations based on r2>0.80 and minor allele frequency ⩾0.05 (Figure 1). All SNPs were genotyped with a custom-by-design 48-Plex SNPscan Kit (Cat#:G0104; Genesky Biotechnologies Inc., Shanghai, China), which was developed according to patented SNP genotyping technology by Genesky Biotechnologies Inc. As presented by Chen et al.,26 it was based on double ligation and multiplex fluorescence PCRs. In order to reduce artefacts due to batch or plate position effects, case and control samples were interspersed within 96-well plates and genotyping was performed blind to case–control status. Furthermore, 78 blind duplicate samples were distributed across all genotyping plates to ensure consistency and each plate included a negative control.
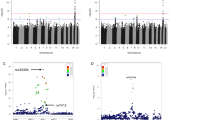
Ten tag SNPs in the genomic region of TOMM40/APOE/APOC1 locus. This track displays transcripts from the Ensembl release 81.38 annotation of the NCBI 38.p3 assembly of the human genome. Each transcript is labeled with both a gene ID and an Ensembl transcript ID. Exons appear as taller shaded areas on the horizontal line that depicts the transcript. Horizontal arrows indicate the transcriptional orientations of individual genes. The scale bar indicated a chromosomal distance of 1.0 kb.
The call rate for each tag SNP was above 98% (Supplementary Table S1). The concordance for duplicate samples was >99%, and only genotypes for SNP rs769450 were out of Hardy–Weinberg equilibrium in all subjects (P=0.0087) and then excluded from further analysis. The 10 tag SNPs successfully genotyped were able to capture 47 common SNPs of the TOMM40/APOE/APOC1 region at r2>0.80 (Supplementary Table S1).
Statistical analysis
Association analysis of TOMM40/APOE/APOC1 locus with human longevity
All analyses were performed using SNPStats (http://bioinfo.iconcologia.net/snpstats/start.htm).27 Hardy–Weinberg equilibrium testing was conducted for all SNPs in all subjects using exact test. To avoid assumptions regarding the modes of inheritance, additive, dominant and recessive models for each polymorphism were assessed. The best-fitting inheritance mode for each polymorphism was determined based on the lowest Akaike information criteria. P-values obtained were corrected (Pc) for multiple testing using Bonferroni for the number of SNPs tested (n=10) and the number of tests performed (n=3).
Haplotype frequencies were estimated using the implementation of the expectation-maximization algorithm. Pairwise r2 was calculated to represent linkage disequilibrium (LD). The association parameters of human longevity were estimated for each haplotype by comparison with a reference haplotype chosen as the most frequent one. Effects associated with rare haplotypes (frequency<0.5%) were determined after categorizing them together. Another web tool SHEsis28 was adopted to yield similar results compared with SNPStats. All P-values presented in this study are two sided, and P<0.05 were considered significant.
In silico identification of evolutionarily conserved regions and transcription factors (TFs) in the flanking regions of longevity-associated SNPs
Because over half of the bases in evolutionarily conserved regions (ECRs) in mammals appear to be functional,29 we examined whether the longevity-associated SNPs and SNPs, which are in strong LD with the longevity-associated SNPs, were in the ECRs. To identify ECRs overlapping these SNPs, we compared the human sequence including these SNPs, with rhesus macaque, dog, mouse, rat, chicken, frog and fugu sequence. Pariwise alignments were produced and the nucleotide level match-mismatch similarity profiles were compared with the blastz algorithm30 incorporated in the ECR browser (http://ecrbrowser.dcode.org/).31, 32 Regions with conserved elements were defined as intervals that exceed the threshold of 100 base pairs with >70% nucleotide identity. The ECRs containing the SNPs were also obtained using the conservation tool of the University of California, Santa Cruz (UCSC) browser (http://genome.ucsc.edu/). Transcription factors (TFs) within the conserved sequences were identified from the Encyclopedia of DNA Elements (ENCODE) chromatin immunoprecipitation (ChIP) followed by high-throughput DNA sequencing (ChIP-seq) data sets at UCSC (Release 2 and 4).
Results
Single-SNP association analysis of TOMM40/APOE/APOC1 locus with human longevity
As Supplementary Table S2 shown, among 10 tag SNPs, rs7254892 in 5′ upstream of TOMM40, three SNPs (rs2075649, rs8106922 and rs1160985) in the intron of TOMM40 and also in 5′ upstream of APOE, as well as rs445925 in 3′ downstream of APOE and also in 5′ upstream of APOC1, were significantly associated with human longevity. For ease of presentation, Table 1 only presents the five significant association polymorphisms in the best mode of inheritance. As shown, the best modes of inheritance were: (1) the dominant effect of SNP rs7254892 (G/A–A/A vs G/G: odds ratio (OR)=1.59, 95% confidence interval (CI)=1.20–2.09, P=0.0011); (2) the recessive effect of SNP rs2075649 (G/G vs A/A–G/A: OR=0.58, 95% CI=0.38–0.87, P=0.0078); (3) the recessive effect of SNP rs8106922 (G/G vs A/A–G/A: OR=0.54, 95% CI=0.34–0.85, P=0.0055); (4) the dominant effect of SNP rs1160985 (C/T-T/T vs C/C: OR=1.29, 95% CI=1.04–1.60, P=0.019); and (5) the dominant effect of SNP rs445925 (G/A–A/A vs G/G: OR=1.40, 95% CI=1.08–1.82, P=0.01). It is suggested that individuals with the minor allele of SNP rs7254892, rs1160985 or rs445925 tended to have longer lifespan than those homozygous for the corresponding major allele, whereas individuals homozygous for the minor allele of rs2075649 or rs8106922 tended to have shorter lifespan than those with the corresponding major allele. However, after adjustment for multiple testing, only one polymorphism (that is, rs7254892) remained significantly associated with human longevity (Pc=0.033; Table 1).
Linkage disequilibrium and haplotype association analysis of TOMM40/APOE/APOC1 locus with human longevity
Pairwise r2 values were lower than 0.80 except those between rs157580 and rs405697 (r2=0.86), as well as between rs405697 and rs439401 (r2=0.81; Figure 2). In the haplotype analysis, three haplotypes were identified to be associated with human longevity (Table 2): compared with the most common haplotype G–G–A–A–C–A–C–A–T–G, both the A–A–A–A–T–A–T–G–C–A and G–A–A–A–T–A–T–G–C–G haplotypes had significantly higher frequency in the cases than in the controls (OR=1.59, 95% CI=1.19–2.12, P=0.0018 and OR=2.02, 95% CI=1.20–3.40, P= 0.0084; respectively), whereas the G–A–G–A–C–G–T–G–T–G haplotype had significantly lower frequency in the cases than in the controls (OR=0.42, 95% CI=0.18–0.94, P=0.035). But after Bonferroni correction, only the A–A–A–A–T–A–T–G–C–A haplotype remained (Pc=0.0216), which suggested that individuals carrying the A–A–A–A–T–A–T–G–C–A haplotype tended to have longer lifespan than the G–G–A–A–C–A–C–A–T–G haplotype carriers.
In silico identification of ECRs and TFs in the flanking regions of longevity-associated SNPs
In the present study, we have observed that one tag SNP (that is, rs7254892) were associated with human longevity after Bonferroni correction. The 1000 Genomes Project database of the phase III CHB and/or CHS populations indicated that five SNPs (rs283808, rs283809, rs283810, rs283813 and rs1160983) are in strong LD with rs7254892 (all pairwise r2>0.800). Therefore, we explored the ECRs and TFs in the regions of chromosome 19: 45386596-45392596 (6001 bp) (including rs283808, rs283809, rs283810, rs283813 and rs7254892) and chromosome 19: 45396729–45397729 (1001 bp) (Human Feb. 2009 (GRCh37/hg19) Assembly) including rs1160983.
SNP rs7254892 and rs283813 were in the ECRs available on both the ECR and UCSC genome browsers (Supplementary Table S3 and Supplementary Figure S1). SNP rs283810 was in the ECRs available on the ECR browser but not on the UCSC genome browser. SNP rs283808 and rs283809 were not in any ECRs. In the UCSC database, several TFs were identified for the ECR containing SNP rs7254892 and/or rs283813, such as signal transducer and activator of transcription 1 (STAT1) and nuclear receptor subfamily 2, group F, member 2 (NR2F2; Supplementary Figure S1). SNP rs1160983 (TOMM40 Exon 6 p.Ser183Ser) was observed in the ECRs (Supplementary Table S4 and Supplementary Figure S2). However, rs1160983 is synonymous and not in the TF-binding sites from the UCSC database (Supplementary Figure S2). The preliminary bioinformatics analysis predicted that SNP rs7254892 or/and rs283813 might affect human longevity by affecting binding regions of TFs.
Discussion
In this study, we conducted research on 10 common tag SNPs across the TOMM40/APOE/APOC1 region to detect novel signatures for association with human longevity in a Chinese population. In initial analysis, five SNPs –rs7254892 in 5′ upstream of TOMM40, three SNPs (rs2075649, rs8106922 and rs1160985) in the intron of TOMM40 and also in 5′ upstream of APOE, as well as rs445925 in 3′ downstream of APOE and also in 5′ upstream of APOC1—showed association with human longevity. But only SNP rs7254892 survived Bonferroni correction. And after Bonferroni correction, only the A–A–A–A–T–A–T–G–C–A haplotype had a significant extension in human lifespan compared with the most common haplotype G–G–A–A–C–A–C–A–T–G.
Four SNPs (rs7254892, rs2075649, rs8106922 and rs1160985) in TOMM40 were found to be significantly associated with human longevity in initial analysis, but only rs7254892 survived Bonferroni correction. SNP rs7254892 has been reported to be associated with atherogenic dyslipidemia in African Americans but not in European Americans at the genome-wide significance threshold.33 SNP rs1160983 (TOMM40 p.Ser183Ser), which is in LD with rs7254892 in the phase III CHB population (r2=0.933, D′=1), also reaches genome-wide significance for association with LDL cholesterol levels in African Americans and European Americans.34 The major allele T of rs283813, which is 422 bp upstream to and in LD with rs7254892 in the phase III CHS population (r2=0.811, D′=1), is genome wide significantly associated with higher LDL cholesterol levels in African Americans.35 Consistently, in the present study, subjects with the minor allele A of rs7254892 tended to have greater life expectancy compared to those without the A allele (that is, those with the G/G genotype; Table 1). The association of SNP rs7254892 with human longevity may possibly be conveyed through its effect on cholesterol metabolism.
SNP rs7254892 is 4881 bp upstream from the transcription start site of TOMM40 and may possibly have an effect on TOMM40 expression. It is also in the intron of poliovirus receptor-related 2 (PVRL2). It or other variants in LD with it may influence the function or expression of PVRL2. However, to date, few functional studies indicated that TOMM40 and PVRL2 participate in cholesterol metabolism. Of note, SNP rs7254892 is also in 5′ upstream of APOE and APOC1 (19 415 and 27 908 bp, respectively, upstream from the transcription start site), which are well-known to be involved in cholesterol metabolism. Bekris et al.36 have demonstrated that four enhancers influence APOE and/or TOMM40 promoter activity according to haplotype and cell type. However, they only focused on exploring the enhancers in the region of chromosome 19: 44888801–44953730 (64.93 kbp, human genome reference assembly GRCh38/hg38), which does not include rs7254892 (chromosome 19: 44886339).
Our preliminary bioinformatics analysis indicated that, among rs7254892 and the five SNPs (rs283808, rs283809, rs283810, rs283813 and rs1160983) which are in strong LD with rs7254892, rs7254892 or/and rs283813 might affect human longevity by affecting binding regions of TFs. More comprehensive efforts and experimental studies will be needed to find the potential functional significance of rs7254892 or other variants linked to it in cholesterol metabolism and life expectancy.
In addition, among the four TOMM40 SNPs, the T allele of rs1160985 has been documented to be genome wide significantly associated with lower LDL levels in Chinese37 and African Americans but not in Hispanic Americans.22 Low LDL levels are a protective factor for cardiovascular disease. Consistently, in this study, subjects with the T allele tended to have longer lifespan compared with those without the T allele (that is, those with the C/C genotype), but this trend did not survive Bonferroni correction (Table 1).
The A allele of rs445925 is genome wide significantly associated with lower LDL levels,38, 39 lower apolipoprotein B and higher APOE,40 as well as lower baseline lipoprotein-associated phospholipase A2 (Lp-PLA2) activity41 in European ancestry individuals. Epidemiological and functional studies have associated elevated Lp-PLA2 enzyme activity with greater risk of developing atherosclerosis and cardiovascular disease, independent from risk associated with circulating lipid levels.42, 43, 44 Taken together, we supposed that SNP rs445925 might be associated with human longevity through its effect on lipid metabolism. Consistently, subjects with the A allele of rs445925 tended to live longer compared with those without the A allele (that is, those with the G/G genotype; Table 1), however, this tendency did not pass the Bonferroni correction.
SNP rs2075650 in TOMM40 have been shown to be significantly associated with human longevity in prior studies,9, 45, 46 but not in the present study. Although there is only moderate LD between rs2075650 and rs429358, rs2075650 is a strong proxy of rs429358 that define APOE ɛ4 (ref. 45) and the association of rs2075650 with longevity is most likely a reflection of the effects of rs429358.9 Rs2075650 and rs445925 are close to and in LD with rs429358 (r2=0.807, D′=0.945) and rs7412 (r2=0.907, D′=1), respectively, in the phase III CHB population. Therefore, we set rs2075650-rs445925 as a proxy for rs429358-rs7412. Rs429358 and rs7412, and their respective differences at amino acids 112 and 158, define the three major isoforms of APOE: ɛ3 (cys112, arg158), ɛ2 (cys112, cys158) and ɛ4 (arg112, arg158). APOE ɛ3, the most common of the three isoforms, is considered to be the normal form. APOE ɛ2 and APOE ɛ4 differ from APOE ɛ3 by single amino-acid substitutions at position 112 or 158. This fourth APOE isoform, which encodes the arg112-cys158 isoform and is referred to as ɛ3r or ɛ1y, are very rare in humans,47 which suggested that rs429358 and rs7412 are two independently associated loci.
The three major isoforms have been widely estimated. Until 26 July 2013, at least 27 studies have examined the relationship between APOE variants and human longevity and/or survival in old age, and 21 of 27 reported a statistically significant association between APOE and longevity (GenAge Database; 2013 http://genomics.senescence.info/genes/longevity.html). Increased APOE ɛ2 frequency and decreased APOE ɛ4 frequency at older ages has been often but not always observed in different populations.6 Therefore, we also conducted haplotype association analysis for SNP rs2075650 and rs445925. As Supplementary Table S5 shown, compared with the A–G haplotype (a proxy for APOE ɛ3), the A–A haplotype (a proxy for APOE ɛ2) was more frequent in LLIs than in younger controls (OR=1.31, 95% CI=1.04–1.67, P=0.024, Pc=0.048), that is, the A–A haplotype was correlated with longer lifespan. Lower G–G haplotype (a proxy for APOE ɛ4) frequency was observed in LLIs, but the trend was not significant (OR=0.82, 95% CI=0.62–1.10, P=0.184, Pc=0.368).
The study included relatively small sample sizes of Li people (n=107). In order to avoid false-positive association owing to population stratification, we also restricted the analysis to those Han people and the results were similar to those in all subjects (Supplementary Tables S6–S8). However, there was one exception—in addition to SNP rs7254892, the recessive effect of rs8106922 on human longevity (G/G vs A/A–G/A: OR=0.48, 95% CI=0.29–0.77, P=0.0016, Pc=0.048) remained in those Han people after Bonferroni correction. It would be of interest to examine these associations in large Li population and replication of these associations in other long-lived populations will be needed to elucidate these results.
Only one haplotype in the TOMM40/APOE/APOC1 region was associated with longer lifespan (A–A–A–A–T–A–T–G–C–A, OR=1.59, 95% CI=1.19–2.12, P=0.0018) when compared with the common referent haplotype after Bonferroni correction. To be noted, this association was similar in magnitude to that observed for the TOMM40 (rs7254892) G/A–A/A variant that was found in 20.3% of LLIs and 13.8% of controls (G/A–A/A vs G/G: OR=1.59, 95% CI=1.20–2.09, P=0.0011), suggesting that this association appeared to be driven by SNP rs7254892.
In conclusion, we have observed that one tag SNP and one haplotype of TOMM40/APOE/APOC1 region were related to human longevity after Bonferroni correction. However, in the absence of functional evidence, the significant SNP or haplotype are not eventually identified as the causal variant or haplotype. The present findings have to be considered as hypothesis generation and will have to be confirmed in further studies.
References
Brooks-Wilson, A. R. Genetics of healthy aging and longevity. Hum. Genet. 132, 1323–1338 (2013).
Panza, F., D'Introno, A., Colacicco, A. M., Capurso, C., Capurso, S., Kehoe, P. G. et al. Vascular genetic factors and human longevity. Mech. Ageing. Dev. 125, 169–178 (2004).
Herskind, A. M., McGue, M., Holm, N. V., Sorensen, T. I., Harvald, B. & Vaupel, J. W. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum. Genet. 97, 319–323 (1996).
Skytthe, A., Pedersen, N. L., Kaprio, J., Stazi, M. A., Hjelmborg, J. V., Iachine, I. et al. Longevity studies in GenomEUtwin. Twin. Res. 6, 448–454 (2003).
Schachter, F., Faure-Delanef, L., Guenot, F., Rouger, H., Froguel, P., Lesueur-Ginot, L. et al. Genetic associations with human longevity at the APOE and ACE loci. Nat. Genet. 6, 29–32 (1994).
Lewis, S. J. & Brunner, E. J. Methodological problems in genetic association studies of longevity—the apolipoprotein E gene as an example. Int. J. Epidemiol. 33, 962–970 (2004).
Soerensen, M., Dato, S., Tan, Q., Thinggaard, M., Kleindorp, R., Beekman, M. et al. Evidence from case-control and longitudinal studies supports associations of genetic variation in APOE, CETP, and IL6 with human longevity. Age (Dordr) 35, 487–500 (2013).
Smith, J. D. Apolipoproteins and aging: emerging mechanisms. Ageing Res. Rev. 1, 345–365 (2002).
Deelen, J., Beekman, M., Uh, H. W., Helmer, Q., Kuningas, M., Christiansen, L. et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell 10, 686–698 (2011).
Nebel, A., Kleindorp, R., Caliebe, A., Nothnagel, M., Blanche, H., Junge, O. et al. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech. Ageing Dev. 132, 324–330 (2011).
Deelen, J., Beekman, M., Uh, H. W., Broer, L., Ayers, K. L., Tan, Q. et al. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum. Mol. Genet. 23, 4420–4432 (2014).
Spinney, L Alzheimer's disease: the forgetting gene. Nature 510, 26–28 (2014).
Helisalmi, S., Hall, A., Merilainen, E. H., Vaisanen, V., Koivisto, A. M., Herukka, S. K. et al. The effect of TOMM40 Poly-T repeat lengths on age of onset and cerebrospinal fluid biomarkers in finnish Alzheimer's disease patients. Neurodegener. Dis. 14, 204–208 (2014).
Devi, L., Prabhu, B. M., Galati, D. F., Avadhani, N. G . & Anandatheerthavarada, H. K. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J. Neurosci. 26, 9057–9068 (2006).
Hansson Petersen, C. A., Alikhani, N., Behbahani, H., Wiehager, B., Pavlov, P. F., Alafuzoff, I. et al. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc. Natl Acad. Sci. USA 105, 13145–13150 (2008).
Abildayeva, K., Berbee, J. F., Blokland, A., Jansen, P. J., Hoek, F. J., Meijer, O. et al. Human apolipoprotein C-I expression in mice impairs learning and memory functions. J. Lipid Res. 49, 856–869 (2008).
Berbee, J. F., Vanmierlo, T., Abildayeva, K., Blokland, A., Jansen, P. J., Lutjohann, D. et al. Apolipoprotein CI knock-out mice display impaired memory functions. J. Alzheimers Dis. 23, 737–747 (2011).
Leduc, V., Jasmin-Belanger, S. & Poirier, J. APOE and cholesterol homeostasis in Alzheimer's disease. Trends Mol. Med. 16, 469–477 (2010).
Vestergaard, M., Hamada, T., Morita, M. & Takagi, M. Cholesterol, lipids, amyloid Beta, and Alzheimer's. Curr. Alzheimer Res. 7, 262–270 (2010).
Keller, M., Schleinitz, D., Forster, J., Tonjes, A., Bottcher, Y., Fischer-Rosinsky, A. et al. THOC5: a novel gene involved in HDL-cholesterol metabolism. J. Lipid Res. 54, 3170–3176 (2013).
Teslovich, T. M., Musunuru, K., Smith, A. V., Edmondson, A. C., Stylianou, I. M., Koseki, M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Coram, M. A., Duan, Q., Hoffmann, T. J., Thornton, T., Knowles, J. W., Johnson, N. A. et al. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet. 92, 904–916 (2013).
Zhou, Q., Zhao, F., Lv, Z. P., Zheng, C. G., Zheng, W. D., Sun, L. et al. Association between APOC1 polymorphism and Alzheimer's disease: a case-control study and meta-analysis. PLoS ONE 9, e87017 (2014).
Beekman, M., Blanche, H., Perola, M., Hervonen, A., Bezrukov, V., Sikora, E. et al. Genome-wide linkage analysis for human longevity: Genetics of Healthy Aging Study. Aging Cell 12, 184–193 (2013).
Fan, X. m [The research of longevity population of the sixth census in China]. Popul. J. 35, 14–20 (2013).
Chen, X., Li, S., Yang, Y., Yang, X., Liu, Y., Liu, Y. et al. Genome-wide association study validation identifies novel loci for atherosclerotic cardiovascular disease. J. Thromb. Haemost. 10, 1508–1514 (2012).
Sole, X., Guino, E., Valls, J., Iniesta, R. & Moreno, V. SNPStats: a web tool for the analysis of association studies. Bioinformatics 22, 1928–1929 (2006).
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z. et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 19, 519–523 (2009).
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigo, R., Gingeras, T. R., Margulies, E. H. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Schwartz, S., Kent, W. J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R. C. et al. Human-mouse alignments with BLASTZ. Genome Res. 13, 103–107 (2003).
Ovcharenko, I., Nobrega, M. A., Loots, G. G. & Stubbs, L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 32, W280–W286 (2004).
Loots, G. & Ovcharenko, I. ECRbase: database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes. Bioinformatics 23, 122–124 (2007).
Avery, C. L., He, Q., North, K. E., Ambite, J. L., Boerwinkle, E., Fornage, M. et al. A phenomics-based strategy identifies loci on APOC1, BRAP, and PLCG1 associated with metabolic syndrome phenotype domains. PLoS Genet. 7, e1002322 (2011).
Lange, L. A., Hu, Y., Zhang, H., Xue, C., Schmidt, E. M., Tang, Z. Z. et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am. J. Hum. Genet. 94, 233–245 (2014).
Elbers, C. C., Guo, Y., Tragante, V., van Iperen, E. P., Lanktree, M. B., Castillo, B. A. et al. Gene-centric meta-analysis of lipid traits in African, East Asian and Hispanic populations. PLoS ONE 7, e50198 (2012).
Bekris, L. M., Lutz, F. & Yu, C. E. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J. Hum. Genet. 57, 18–25 (2012).
Zhou, L., He, M., Mo, Z., Wu, C., Yang, H., Yu, D. et al. A genome wide association study identifies common variants associated with lipid levels in the Chinese population. PLoS ONE 8, e82420 (2013).
Smith, E. N., Chen, W., Kahonen, M., Kettunen, J., Lehtimaki, T., Peltonen, L. et al. Longitudinal genome-wide association of cardiovascular disease risk factors in the Bogalusa heart study. PLoS Genet. 6, e1001094 (2010).
Trompet, S., de Craen, A. J., Postmus, I., Ford, I., Sattar, N., Caslake, M et al. Replication of LDL GWAs hits in PROSPER/PHASE as validation for future (pharmaco)genetic analyses. BMC Med. Genet. 12, 131 (2011).
Surakka, I., Whitfield, J. B., Perola, M., Visscher, P. M., Montgomery, G. W., Falchi, M. et al. A genome-wide association study of monozygotic twin-pairs suggests a locus related to variability of serum high-density lipoprotein cholesterol. Twin Res. Hum. Genet. 15, 691–699 (2012).
Chu, A. Y., Guilianini, F., Grallert, H., Dupuis, J., Ballantyne, C. M., Barratt, B. J. et al. Genome-wide association study evaluating lipoprotein-associated phospholipase A2 mass and activity at baseline and after rosuvastatin therapy. Circ. Cardiovasc. Genet. 5, 676–685 (2012).
Thompson, A., Gao, P., Orfei, L., Watson, S., Di Angelantonio, E., Kaptoge, S. et al. Lipoprotein-associated phospholipase A(2) and risk of coronary disease, stroke, and mortality: collaborative analysis of 32 prospective studies. Lancet 375, 1536–1544 (2010).
Hakkinen, T., Luoma, J. S., Hiltunen, M. O., Macphee, C. H., Milliner, K. J., Patel, L. et al. Lipoprotein-associated phospholipase A(2), platelet-activating factor acetylhydrolase, is expressed by macrophages in human and rabbit atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 19, 2909–2917 (1999).
Kolodgie, F. D., Burke, A. P., Skorija, K. S., Ladich, E., Kutys, R., Makuria, A. T. et al. Lipoprotein-associated phospholipase A2 protein expression in the natural progression of human coronary atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 26, 2523–2529 (2006).
Sebastiani, P., Solovieff, N., Dewan, A. T., Walsh, K. M., Puca, A., Hartley, S. W. et al. Genetic signatures of exceptional longevity in humans. PLoS ONE 7, e29848 (2012).
Lu, F., Guan, H., Gong, B., Liu, X., Zhu, R., Wang, Y. et al. Genetic variants in PVRL2-TOMM40-APOE region are associated with human longevity in a Han Chinese population. PLoS ONE 9, e99580 (2014).
Seripa, D., Matera, M. G., Daniele, A., Bizzarro, A., Rinaldi, M., Gravina, C. et al. The missing ApoE allele. Ann. Hum. Genet. 71, 496–500 (2007).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No.31460290), Department of Science and Technology of Hainan Province (KJHZ2013-16 and ZDXM20090805), the Research Start-Up Fund in Hainan Medical College for Dr Lin, the Young Scientists Fund of the National Natural Science Foundation of China (Grant No.31100904), Fund for Less Developed Regions of the National Natural Science Foundation of China (Grant No. 81460184). In addition, we thank Dr Zhengwen Jiang, Dr Yan Liu and Dr Ying Wang (Genesky Biotechnologies Inc., Shanghai, China) for technical support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Lin, R., Zhang, Y., Yan, D. et al. Association of common variants in TOMM40/APOE/APOC1 region with human longevity in a Chinese population. J Hum Genet 61, 323–328 (2016). https://doi.org/10.1038/jhg.2015.150
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.150
This article is cited by
-
Heterogeneity in Alzheimer's Disease Diagnosis and Progression Rates: Implications for Therapeutic Trials
Neurotherapeutics (2022)
-
Effects of the association between APOE rs405509 polymorphisms and gene-environment interactions on hand grip strength among middle-aged and elderly people in a rural population in southern China
Journal of Orthopaedic Surgery and Research (2021)
-
Mining Outcome-relevant Brain Imaging Genetic Associations via Three-way Sparse Canonical Correlation Analysis in Alzheimer’s Disease
Scientific Reports (2017)