Abstract
Recently, de novo KIF1A mutations were identified in patients with intellectual disability, spasticity and cerebellar atrophy and/or optic nerve atrophy. In this study, we analyzed a total of 62 families, including 68 patients with genetically unsolved childhood cerebellar atrophy, by whole-exome sequencing (WES). We identified five de novo missense KIF1A mutations, including only one previously reported mutation (p.Arg316Trp). All the mutations are located in the motor domain of KIF1A. In all patients, initial symptom onset was during the infantile period, and included developmental delay in three patients and gait disturbance in two. Thereafter, they showed gait disturbances, exaggerated deep tendon reflexes, cerebellar symptoms and cerebellar atrophy on brain magnetic resonance imaging. Four patients showed lower limb spasticity, upper limb clumsiness and visual disturbances. Nerve conduction study revealed peripheral neuropathy in three patients. This study further delineates clinical features of de novo KIF1A mutations. Genetic testing of KIF1A should be considered in children with developmental delay, cerebellar atrophy and pyramidal features.
Similar content being viewed by others
Introduction
Kinesin superfamily proteins (KIFs) are motor proteins that play important roles in transport of various cargos along microtubules.1, 2 KIFs comprise three major groups depending on the position of the motor domain: N-terminal motor domain KIFs, middle motor domain KIFs and C-terminal motor domain KIFs.1 KIF1A belongs to the N-terminal motor domain KIFs and is composed of a motor domain, stark domain and tail region.1 The motor domain binds to microtubules and moves along them by hydrolyzing adenosine triphosphate (ATP), whereas the tail region recognizes and binds to the cargo.1
Previous reports have identified KIF1A recessive mutations in patients with hereditary sensory and autonomic neuropathy type 2, and hereditary spastic paraplegias (SPG30).3, 4, 5 Recently, de novo KIF1A mutations were identified in patients with intellectual disability, spasticity and cerebellar atrophy and/or optic nerve atrophy6, 7, 8 that overlap with clinical features caused by recessive KIF1A mutations. Therefore, abnormal KIF1A function can affect both the central and peripheral nervous systems.
Here, we performed whole-exome sequencing (WES) of patients with childhood cerebellar atrophy. We identified de novo KIF1A mutations in five patients and analyzed their clinical phenotypes.
Patients and Methods
Patients
We have previously described WES analysis of 25 patients with cerebellar atrophy.9 In this study, a total of 62 families (including 68 patients with childhood cerebellar atrophy) were newly recruited as a second cohort and analyzed by WES. From the clinical point of view, it is difficult to make a definite genetic diagnosis in each patient. Both static and progressive cerebellar atrophy were included. Detailed clinical information was obtained from the clinicians.
Genetic analysis
Genomic DNA was obtained from peripheral blood leukocytes using QuickGene 610L (Wako, Osaka, Japan). Genomic DNA was captured using the SureSelect Human All Exon v4 or v5 Kit (51 Mb; Agilent Technologies, Santa Clara, CA, USA) and sequenced on a HiSeq2000 (Illumina, San Diego, CA, USA) with 101-bp paired-end reads. Exome data processing, variant calling and variant annotation were performed as described previously.10 Rare nonsynonymous KIF1A variants, which were absent in dbSNP 137, the 6500 exomes of the National Heart, Lung and Blood Institute exome project, and our in-house 575 control exomes, were considered as candidate KIF1A mutations, and their segregation was examined by Sanger sequencing with trio samples (patients and their parents). In families showing de novo mutations, parentage was confirmed by microsatellite analysis, as previously described.11 Pathogenicity of the mutations was predicted using Sorting Intolerant from Tolerant (SIFT; http://sift.jcvi.org/), Polyphen2 (http://genetics.bwh.harvard.edu/pph2/) and Mutation Taster (http://www.mutationtaster.org/). KIF1A mutations were annotated based on transcript variant 1 (NM_001244008.1). The de novo KIF1A mutations were deposited to a gene-specific database (http://databases.lovd.nl/shared/genes/KIF1A).
Standard protocol approvals and patient consents
Experimental protocols were approved by the institutional review board of Yokohama City University School of Medicine. Written informed consent was obtained from all individuals and/or their families in compliance with relevant Japanese regulations.
Results and discussion
WES yielded an average of 87.1 million reads per sample (range 47.6–164.7 million reads per sample), resulting in an average read depth of 104.3 on the all RefSeq coding sequence (build 37/hg 19, range across all samples: 56.6–192.8). A total of five candidate missense KIF1A mutations were found in five patients, and all were confirmed as de novo events by Sanger sequencing using trio samples. In these five patients, no other candidate mutations, which were consistent with genetic model and segregation, were found in the other 180 genes previously reported in cerebellar atrophy.9 All mutations were located in the motor domain (5/5, 100%) and substituted evolutionarily conserved amino acids (Figure 1). One mutation (p.Arg316Trp) had been previously reported.7 SIFT, Polyphen2 and Mutation Taster predicted that all the mutations are highly damaging to the structure of KIF1A (Supplementary Table S1).
Schematic presentation of KIF1A and evolutionary conservation of substituted amino acids by KIF1A mutations. The KIF1A subunit contains three domains: motor domain, forkhead-associated domain (FHA) and pleckstrin homology domain (PH).7 Mutations are annotated according to NM_001244008.1. The p.Arg316Trp mutation was reported previously (red characters).7 All the KIF1A mutations occur at evolutionarily conserved amino acids. Orthologous sequences were aligned using CLUSTALW (http://www.genome.jp/tools/clustalw/). A full color version of this figure is available at the Journal of Human Genetics journal online.
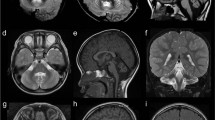
Clinical information on the patients with KIF1A mutations are summarized in Table 1, and magnetic resonance imaging findings are shown in Figure 2. Initial symptom onset was during the infantile period in all patients, with developmental delay in 3 patients (3/5, 60%) and gait disturbance in 2 patients (2/5, 40%). Subsequently, all patients showed gait disturbances, exaggerated deep tendon reflexes, cerebellar symptoms and cerebellar atrophy on brain magnetic resonance imaging (5/5, 100%). Four patients showed lower limb spasticity (4/5, 80%) and one patient had hypotonia (1/5, 20%). Three patients showed peripheral neuropathy that was demonstrated by abnormal nerve conduction studies (3/5, 60%). Three patients showed muscle weakness (3/5, 60%), and two had muscle hypertrophy (2/5, 40%). Upper limb clumsiness was observed in four patients (4/5, 80%). Three patients showed optic nerve atrophy (3/5, 60%), whereas one patient had hypermetropic astigmatism and light amblyopia (1/5, 20%). In patients 2 and 3, cerebellar atrophy was more severe in the vermis than the hemisphere. Periventricular white matter hyperintensities were observed on fluid-attenuated inversion recovery images in two patients (2/5, 40%). Dentate nucleus hyperintensity was observed on T2-weighted images in one patient (1/5, 20%). Case reports are available in Supplementary Information.
Brain magnetic resonance imaging (MRI) of patients. (a, j) T2-weighted axial images, (b) T2-weighted coronal image, (c, d, f, i) T1-weighted sagittal images, (e) fluid-attenuated inversion recovery (FLAIR) coronal and (g) axial images and (h) T1-weighted coronal image. (a, b) Patient 1 at 6 years of age, (c) patient 2 at 11 years, (d, e) patient 3 at 6 years, (f, g) patient 4 at 28 years and (h, i, j) patient 5 at 8 years. Cerebellar atrophy was observed in all patients (a−f, h, i). In patient 4, a slightly high intensity area surrounding the lateral ventricles bilaterally was observed on FLAIR images (arrows). In patient 5, a hyperintense dentate nucleus was observed on T2-weighted images (arrowheads).
All the de novo KIF1A mutations we have identified here are located in the motor domain, and predicted to affect motor function based on structural models. Arg254, and Arg307 and Arg316 are located on loop L11 and the α5 helix of the switch II cluster, respectively, that associates γ-phosphate release during ATP hydrolysis and KIF1A binding to microtubules.7, 12, 13 The p.Glu253Lys mutation adjacent to Arg254 was previously predicted to change the charge of salt bridge-forming residues, resulting in suppression of γ-phosphate release.7 In addition, the p.Arg316Trp mutation was predicted to disrupt stabilization of loop L8 that binds to microtubules.7, 13 Thus, it is likely that the three mutations we have identified disrupt function of the switch II cluster. Located on loop L7, Glu148 is a Mg-stabilizer along with Arg203 and Asp248.14 Therefore, the Glu148 mutation may affect Mg stability that is crucial for kinesin regulation and mortality.14
The clinical features of the five patients reported here are consistent with previous reports in which patients with de novo KIF1A mutations show intellectual disability, peripheral neuropathy, cerebellar atrophy, optic nerve atrophy and lower limb spasticity.6, 7, 8 It is interesting to note that two of our patients showed periventricular white matter hyperintensities on fluid-attenuated inversion recovery images, and one patient showed dentate nucleus hyperintensity on T2-weighted images.
In conclusion, we have identified five de novo KIF1A mutations in patients with childhood cerebellar atrophy. Our data shed light on understanding the phenotypic spectrum of de novo KIF1A mutations.
References
Hirokawa, N., Niwa, S. & Tanaka, Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 (2010).
Hirokawa, N., Noda, Y., Tanaka, Y. & Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 (2009).
Klebe, S., Azzedine, H., Durr, A., Bastien, P., Bouslam, N., Elleuch, N. et al. Autosomal recessive spastic paraplegia (SPG30) with mild ataxia and sensory neuropathy maps to chromosome 2q37.3. Brain 129, 1456–1462 (2006).
Klebe, S., Lossos, A., Azzedine, H., Mundwiller, E., Sheffer, R., Gaussen, M. et al. KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur. J. Hum. Genet. 20, 645–649 (2012).
Riviere, J. B., Ramalingam, S., Lavastre, V., Shekarabi, M., Holbert, S., Lafontaine, J. et al. KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am. J. Hum. Genet. 89, 219–230 (2011).
Hamdan, F. F., Gauthier, J., Araki, Y., Lin, D. T., Yoshizawa, Y., Higashi, K. et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am. J. Hum. Genet. 88, 306–316 (2011).
Lee, J. R., Srour, M., Kim, D., Hamdan, F. F., Lim, S. H., Brunel-Guitton, C. et al. De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy and cerebellar atrophy. Hum. Mutat. 36, 69–78 (2015).
Okamoto, N., Miya, F., Tsunoda, T., Yanagihara, K., Kato, M., Saitoh, S. et al. KIF1A mutation in a patient with progressive neurodegeneration. J. Hum. Genet. 59, 639–641 (2014).
Ohba, C., Osaka, H., Iai, M., Yamashita, S., Suzuki, Y., Aida, N. et al. Diagnostic utility of whole exome sequencing in patients showing cerebellar and/or vermis atrophy in childhood. Neurogenetics 14, 225–232 (2013).
Saitsu, H., Nishimura, T., Muramatsu, K., Kodera, H., Kumada, S., Sugai, K. et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 45, 445–449, 449e1 (2013).
Saitsu, H., Kato, M., Mizuguchi, T., Hamada, K., Osaka, H., Tohyama, J. et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat. Genet. 40, 782–788 (2008).
Kikkawa, M., Sablin, E. P., Okada, Y., Yajima, H., Fletterick, R. J. & Hirokawa, N. Switch-based mechanism of kinesin motors. Nature 411, 439–445 (2001).
Nitta, R., Kikkawa, M., Okada, Y. & Hirokawa, N. KIF1A alternately uses two loops to bind microtubules. Science 305, 678–683 (2004).
Nitta, R., Okada, Y. & Hirokawa, N. Structural model for strain-dependent microtubule activation of Mg-ADP release from kinesin. Nat. Struct. Mol. Biol. 15, 1067–1075 (2008).
Acknowledgements
We are grateful to the patients and their families for their participation in this study. We thank Nobuko Watanabe, Kiyomi Masuko, Sayaka Sasamoto and Mai Satoh for their excellent technical assistance. This work was supported by the Ministry of Health, Labour and Welfare of Japan; the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research (B) (25293085, 25293235), Grant-in-Aid for Challenging Exploratory Research (26670505), Grant-in-Aid for Scientific Research (A) (13313587) and Grant-in-Aid for Scientific Research (C) (26461549, 24591500, 26330331)); the Takeda Science Foundation; the Fund for Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program in the Project for Developing Innovation Systems from the Japan Science and Technology Agency; the Strategic Research Program for Brain Sciences (11105137); and a Grant-in-Aid for Scientific Research on Innovative Areas (Transcription Cycle) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (12024421).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Ohba, C., Haginoya, K., Osaka, H. et al. De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. J Hum Genet 60, 739–742 (2015). https://doi.org/10.1038/jhg.2015.108
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.108
This article is cited by
-
Monoallelic KIF1A-related disorders: a multicenter cross sectional study and systematic literature review
Journal of Neurology (2022)
-
Challenges and Controversies in the Genetic Diagnosis of Hereditary Spastic Paraplegia
Current Neurology and Neuroscience Reports (2021)
-
KIF1A-related autosomal dominant spastic paraplegias (SPG30) in Russian families
BMC Neurology (2020)
-
KIF1A variants are a frequent cause of autosomal dominant hereditary spastic paraplegia
European Journal of Human Genetics (2020)
-
Mutations in the KIF21B kinesin gene cause neurodevelopmental disorders through imbalanced canonical motor activity
Nature Communications (2020)