Abstract
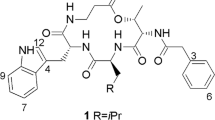
A new cyclic depsipeptide, rakicidin F (1), along with the known compound rakicidin C (2), was isolated from the fermentation broth of the marine sponge-derived actinomycete strain Streptomyces sp. GKU 220. Their structures were elucidated by interpreting the HRFABMS and NMR spectroscopic data. Rakicidin F (1) showed growth inhibitory activity against bacteria.
Similar content being viewed by others
Main
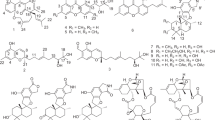
Marine sponges are known to harbor diverse microbial communities.1 Among these communities, actinomycetes are considered to be one of the most promising resources for new bioactive compounds of value in human/veterinary medicine and agriculture.2 We previously reported a new rakicidin derivative that was isolated from the marine sediment-derived Streptomyces sp. MWW064 and shown to inhibit tumor cell invasion.3 In the course of our continuing screening program to discover structurally unique secondary metabolites from actinomycetes isolated from different ecosystems in Thailand,4, 5, 6, 7 a new antibacterial cyclic depsipeptide, rakicidin F (1), along with the known compound rakicidin C (2) (Figure 1a),8 was isolated from the fermentation broth of the marine sponge-derived actinomycete strain Streptomyces sp. GKU 220. Herein, we report the fermentation, isolation, structural elucidation and biological activities of compounds 1 and 2.
The producing organism, GKU 220, was isolated from a marine sponge sample collected in Andaman sea, Ranong, Thailand. The morphological characteristics and 16 S rRNA gene sequence demonstrated that the strain belongs to the genus Streptomyces. Spores of the strain were inoculated into 20 ml of f medium consisting of casitone 0.75%, yeast extract 0.75%, glycerol 1.5% and NaCl 0.25% with no pH adjustment in a 100-ml Erlenmeyer flask, and cultured on a reciprocating shaker at 28 °C for 3 days (120 s.p.m.). The seed culture (1 ml) was inoculated in a 500-ml baffled flask containing A-16 medium (glucose 2%, Pharmamedia 1% and CaCO3 0.5% with no pH adjustment) as the production medium. Fermentation was carried out at 28 °C for 7 days under shaking conditions (120 r.p.m.), after which the culture (3 l) was separated by centrifugation. The culture supernatant was extracted with an equal volume of ethyl acetate (EtOAc) three times and the mycelium was extracted with EtOH. The EtOAc and EtOH-soluble portions were concentrated under reduced pressure, and then the two portions were combined to yield 1.1 g crude extract. The crude extract was separated into seven fractions by silica gel column chromatography (CHCl3-MeOH, gradient 1:0-0:1 (v/v)). Fraction 4 (CHCl3-MeOH, 4:1 (v/v)) was further purified by a reversed-phase HPLC system on a Capcell-Pak C18 column (UG 80; 5 μm, 10 i.d. x 250 mm; Shiseido, Tokyo, Japan) developed with isocratic elution (38% CH3CN at 5 ml min−1 and UV detection at 254 nm) to yield pure compounds 1 (5.1 mg) eluting at 20.4 min and 2 (7.9 mg) eluting at 13 min.
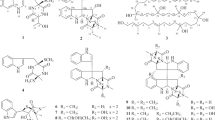
Compounds 1 and 2 were obtained as a white amorphous powder. The molecular formula of 1 was deduced to be C27H44N4O6 by HRFABMS (m/z 521.3345 [M+H]+, calcd for C27H45N4O6, 521.3339). The molecular formula of 2 was also determined to be C26H42N4O6 in the same manner (m/z 507.3196 [M+H]+, calcd for C26H43N4O6, 507.3183). The IR absorption bands of compound 1 suggested the presence of OH or NH (3372 cm−1) and carbonyl (1655 and 1616 cm−1) functionalities. The 1H and 13C NMR spectra of 1 in conjunction with DEPT-135 and HSQC spectrum presented six methyls at δC 37.8, 20.7, 20.7, 16.0 14.4 and 11.7, seven methylenes at δC 120.5, 54.1, 46.6, 43.3, 33.1, 31.0 and 30.0, and eight methines at δC 141.4, 119.0, 78.1, 53.9, 44.3, 32.9, 32.2 and 28.3 (Table 1). In addition, six quaternary carbon atoms were observed at δC 177.3, 175.9, 171.1, 170.3, 168.9 and 139.5, including five carbonyl carbons at δC 177.3, 175.9, 171.1, 170.3 and 168.9, and one olefinic quaternary carbon at δC 139.5. The 1H-1H COSY correlations revealed three partial structures: H-2 to H-4, H-10 to H-11, and H-15 to H-23, including four methyl substitutions (Figure 1b). Two olefinic methylene protons (δH 5.54, 5.46, H2-13) showed HMBC correlations to an olefinic carbon C-11 (δC 141.4) and a quaternary carbon C-12 (δC 139.5), while a methine proton (δH 6.20) at C-10 coupled to a carbonyl C-9 carbon (δC 168.9), which revealed the partial structure (C-9 to C-13) in the 2,4-pentadienoate moiety. The geometry of the double bond between C-10 and C-11 was assigned as E on the basis of the vicinal coupling constant (J10,11=14.7 Hz in average). The 1H-13C long-range correlations from H-8 (δH 3.13) to C-9 and C-7 (δC 54.1), and from H-7 (δH 4.47, 3.99) to C-9 and a carbonyl carbon C-6 (δC 170.3) confirmed a connection between the 2,4-pentadienoate moiety (C-9 to C-13) and N-methylglycine moiety. The HMBC correlations from methylene protons (δH 2.29, H2-4) to a carbonyl carbon C-5 (δC177.3) and from methyl protons (δH 1.22, H3-27) to a carbonyl carbon C-14 (δC 175.9) revealed that the carbonyl carbons were located at positions adjacent to C-4 and C-15, respectively. These interpretations indicated that compound 1 has three partial structures, as described above, and the chemical shifts showed high similarity to those of rakicidins A, B, C, D and E.3, 8, 9, 10
The structure of 1 was finally established by comparison with the NMR spectra of 2. The chemical shifts of 1 and 2 were almost identical except for a methyl and a methylene residue in the acyl group (C-14 to C-27 in 1) (Supplementary Table 1). The chemical shifts of a methyl (δC 11.7 δH 0.88) at C-23 and a methylene (δC 30.0 δH 1.42, 1.07) at C-22 were observed in the NMR spectra of 1, whereas a new methyl signal (δC 24.5 δH 0.89) at C-22 came along in the NMR spectra of 2. Taken together with the difference of the predicted molecular formula between 1 and 2, these findings suggested that compound 2 is a one-methylene-shorter derivative of 1. HMBC correlations of 2 from a methine proton (δH 4.59, H-2) to a carbonyl carbon C-6 (δC 170.4) and a carbonyl carbon C-1 (δC 171.1) and from a methine proton (δH 5.38, H-16) to C-1 confirmed that there was a connection among three partial fragments: C-1 to C-5, C-6 to C-13, and C-15 to C-26. In addition, the proton and carbon chemical shifts were almost identical with those of rakicidin C as reported in the literature.8 Thus, we concluded that compound 2 is rakicidin C and that the structure of rakicidin F (1) is a one-methylene-longer derivative of rakicidin C (2) (Figure 1a).
Compounds 1 and 2 were evaluated for their antimicrobial activity against Bacillus subtilis, Escherichia coli and Saccharomyces cerevisiae by a paper disk diffusion assay. Rakicidin C has been reported to be inactive against Gram-positive and Gram-negative strains and has no cytotoxicity toward various cell lines, although the details of the strains, cell lines and experimental methods were not provided in the literature.8 Rakicidin F (1) showed growth inhibitory activity against B. subtilis and E. coli at the dosage of 25 μg per disk, while rakicidin C (2) showed weak activity only against B. subtilis with an MIC value of 50 μg per disk. Rakicidin C (2) treated at a dose of 1.25 μM achieved 30% inhibition of invading murine carcinoma colon 26-L5 cells without any cytotoxicity,11 and rakicidin F (1) did not show any anti-invasive effect at non-cytotoxic concentration. Meanwhile, rakicidin C (2) is less cytotoxic than rakicidin F (1) (IC50=17 μM).
In conclusion, marine sponge-derived Streptomyces sp. GKU 220 produces a novel antibacterial cyclic depsipeptide rakicidin F (1) consisting of three amino acids, including a rare and nonproteinogenic amino acid (4-amino-2,4-pentadienoic acid), and a modified fatty acid. Rakicidins F (1) and C (2) have a different 3-hydroxy acid moiety from rakicidins A (3), B (4), D (5) and E (6), and a glutamine residue in place of the 3-hydroxyasparagine residue in 3, 4 and 5 (Figure 1a).3, 8, 9, 10 Further biological and structure-activity relationship studies could expand the utility of rakicidins in the development of anti-tumor agents.
Dedication
The authors dedicate this work to Professor Hamao Umezawa who made significant contributions to the discovery of antibiotics, especially including kanamycin, used in human medicine and agriculture.
References
Hentschel, U., Piel, J., Degnan, S. M. & Taylor, M. W. Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 10, 641–654 (2012).
Abdelmohsen., U. R., Bayer, K. & Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 31, 381–399 (2014).
Igarashi, Y. et al. Rakicidin D, an inhibitor of tumor cell invasion from marine-derived Streptomyces sp. J. Antibiot. 63, 563–565 (2010).
Yu, L. et al. Jomthonic acids B and C, two new modified amino acids from Streptomyces sp. J. Antibiot. 67, 345–347 (2014).
Siriwach, R. et al. Bipolamides A and B, triene amides isolated from the endophytic fungus Bipolaris sp. MU34. J. Antibiot. 67, 167–170 (2014).
Igarashi, Y. et al. Jomthonic acid, a modified amino acid from a soil-derived Streptomyces. J. Nat. Prod. 75, 986–990 (2012).
Igarashi, Y. et al. Prajinamide, a new modified peptide from a soil-derived Streptomyces. J. Antibiot. 65, 157–159 (2012).
Hu, J.-F. et al. Rakicidin C, a new cyclic depsipeptide from Streptomyces sp. Eur. J. Org. Chem. 3353–3356 (2000).
McBrien, K. D. et al. Rakicidins, new cytotoxic lipopeptides from Micromonospora sp. Fermentation, isolation and characterization. J. Antibiot. 48, 1446–1452 (1995).
Oku, N. et al. Complete stereochemistry and preliminary structure-activity relationship of rakicidin A, a hypoxia-selective cytotoxin from Micromonospora sp. J. Nat. Prod. 77, 2561–2565 (2014).
Saito, K. I. et al. A modified and convenient method for assessing tumor cell invasion and migration and its application to screening for inhibitors. Biol. Pharm. 20, 345–348 (1997).
Acknowledgements
This work was supported in part by a Grant-in-Aid for Scientific Research (C) (Grant Number JP15K07358) from the Japan Society for the Promotion of Science (JSPS) to SK; by a grant from the Salt Science Research Foundation (No. 1117) to SK; and by a grant for the Joint Program in the Field of Biotechnology from JSPS, the National Research Council of Thailand, and the National Science and Technology Development Agency of Thailand to TN and YI.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on The Journal of Antibiotics website
Supplementary information
Rights and permissions
About this article
Cite this article
Kitani, S., Ueguchi, T., Igarashi, Y. et al. Rakicidin F, a new antibacterial cyclic depsipeptide from a marine sponge-derived Streptomyces sp.. J Antibiot 71, 139–141 (2018). https://doi.org/10.1038/ja.2017.92
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2017.92
This article is cited by
-
Development of a drug discovery approach from microbes with a special focus on isolation sources and taxonomy
The Journal of Antibiotics (2023)