Abstract
A new depsidone, named 7-chlorofolipastatin, and five known structurally related depsidones were isolated from the culture broth of the marine-derived fungus Aspergillus ungui NKM-007 by solvent extraction and HPLC using an octadecylsilyl column. The structure of 7-chlorofolipastatin was elucidated by various spectroscopic data including 1D and 2D NMR spectroscopy. 7-Chlorofolipastatin inhibited sterol O-acyltransferase (SOAT) 1 and 2 isozymes in cell-based and enzyme assays using SOAT1- and SOAT2-expressing Chinese hamster ovary (CHO) cells.
Similar content being viewed by others
Introduction
The enzyme sterol O-acyltransferase (SOAT, also known as acyl-coenzyme A:cholesterol acyltransferase), an endoplasmic reticulum membrane protein, plays important roles in cholesterol regulation in humans.1 SOAT may be a promising target for the development of new anti-atherosclerotic agents. Although pharmaceutical laboratories have identified a number of synthetic SOAT inhibitors, none have been successfully developed to date because of side effects or the lack/absence of efficacy in clinical trials.2, 3, 4, 5 Recent molecular biological studies revealed the existence of two SOAT isozymes, SOAT1 and SOAT2, with distinct functions.6, 7, 8, 9 SOAT1 is ubiquitously expressed in all tissues and cells, whereas SOAT2 is expressed predominantly in the liver (hepatocytes) and intestine.10
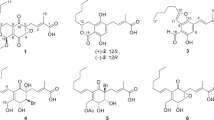
Our group has an extensive experience in discovering SOAT inhibitors with a microbial origin in an enzyme assay using rat liver microsomes.11. After confirming the presence of two SOAT isozymes, we reexamined SOAT inhibitors in cell-based and enzyme assays utilizing Chinese hamster ovary (CHO) cells expressing African green monkey SOAT1 (SOAT1-CHO cells) and SOAT2 (SOAT2-CHO cells) to investigate the selectivity of inhibitors toward these isozymes.12, 13 We have continued this screening program in consideration of the selectivity of candidate microbial cultures toward the SOAT isozymes in these assay systems. A new chlorinated depsidone, designated 7-chlorofolipastatin (1; Figure 1), was recently isolated along with five known depsidones: folipostatin (2),14 unguinol (3),15 2-chlorounginol (4),16 2,7-dichlorounginol (5)17 and nornidulin (6)18 from the culture broth of the marine-derived fungus Aspergillus ungui NKH-007. The fermentation, isolation, structural elucidation and SOAT inhibitory activities of 1 to 6 have been described in the present study.
Results and discussion
Isolation of 1 to 6
The culture broth (0.6 l) was treated with EtOH (0.6 l) for 1 h, and EtOH extracts were filtered to remove cell debris. After the concentration of extracts to remove EtOH, the aqueous solution was adjusted to pH 9.0 and extracted with EtOAc (0.6 l). The organic layer was dried over Na2SO4 and concentrated under reduced pressure to give brown materials (307 mg). These materials were dissolved in a small amount of MeOH and purified by HPLC under the following conditions: column, CAPCELL PAK C18 (i.d. 20 × 250 mm; Shiseido, Tokyo, Japan); mobile phase, 50% CH3CN containing 0.05% H3PO4; flow rate, 8 ml min−1; detection, UV 210 nm. Compounds 1 to 6 were eluted a peak with retention times of 78, 41, 22, 34, 55 and 83 min, respectively. Each fraction of the peak was collected and concentrated to remove CH3CN. The aqueous solution was extracted with EtOAc and the organic layer was concentrated to dryness to give pure 1 (27.3 mg), 2 (19.3 mg), 3 (33.0 mg), 4 (26.0 mg), 5 (23.4 mg) and 6 (15.7 mg) as white powders.
Structural elucidation of 1
The physicochemical properties of 1 are summarized in Table 1. In UV spectra, 1 showed absorption maxima at 226 and 278 nm in MeOH. Broad OH absorption near 3414 cm−1, typical C-H (CH2) stretching absorptions at 2924 and 2857 cm−1, carbonyl absorption at 1711 cm−1 and aromatic C-C stretch absorptions (for carbon–carbon bonds in the aromatic ring) at 1600 and 1412 cm−1 were observed in IR spectra. The electrospray ionization (EI)-MS of 1 showed an isotope pattern of m/z 413 [M-H]− and 415 [M+2-H]− at a ratio of 3:1, indicating the presence of one chlorine atom in the molecule. The molecular formula C23H23O5Cl was assigned on the basis of its EI-MS (m/z 437.1134 [M+Na]+, Δ−0.3 mmu), indicating 12 degrees of unsaturation.
The 1H and 13C NMR spectra of 1 in DMSO-d6 (Table 2) showed 23 proton and 23 carbon signals that were confirmed by an analysis of 2D NMR correlations. The multiplicity of the carbon signals was classified into five methyl carbons, three sp2 methine carbons, one sp2 nitrogenated methine carbon, eight sp2 quaternary carbons, five sp2 oxygenated quaternary carbons and one carbonyl carbon by an analysis of distorsionless enhancement by polarization transfer (DEPT) and heteronuclear multiple-quantum correlation (HMQC) data. The connectivity of proton and carbon atoms was established by HMQC(Table 2). As shown in Figure 2, the partial structures I and II were elucidated by 1H–1H correlation spectroscopy (COSY) spectra. The 13C–1H long-range couplings of 2J and 3J in the heteronuclear multiple bond correlation (HMBC) spectra (Figure 2) proved the presence of the following linkages. (1) Cross peaks from the sp2 methine proton H-2 (δ 6.55) to the sp2 quaternary carbon C-3 (δ 159.9), the sp2 quaternary carbon C-4 (δ 113.3) and the sp2 carbon C-11a (δ 110.0), from the hydroxyl proton OH-3 (δ 10.54) to C-3 and the sp2 carbon C-4 (δ 113.3) and from the methyl proton 4-CH3 (δ 2.04) to C-3, C-4 and the sp2 oxygenated carbon C-4a (δ 161.3) suggested the presence of a 3-hydroxy-4-methyl-1,4a,11a-pentasubtituted benzene ring. Further long-range couplings from the sp2 methine proton H-2′ (δ 5.34) to the methyl carbon C-4′(δ 17.5), from the methyl proton H3-3′ (δ 1.64) to the sp2 carbon C-1′ (δ 135.7) and from the methyl proton H3-4′ (δ 1.77) to C-1′ and the sp2 methine carbon C-2′ (δ 123.9) suggested the presence of an isobutyl group containing the partial structure I. This isobutyl group was connected to the benzene ring at C-1 by an observation of the 3J cross peaks from H-2′ and H3-4′ to the carbon C-1 (δ 148.4) and of the 4J cross peak from H-3′to C-1 in HMBC experiments. Furthermore, a 4J cross peak was observed from H-2 to the carbonyl carbon C-11 (δ 163.0) in HMBC experiments, suggesting that this carbonyl carbon was connected to the benzene ring at C-11a. Thus, the ring A of 1 was elucidated as shown in Figure 2. (2) The ring B of 1 was elucidated from an analysis of the long-range couplings in HMBC in the same manner as described above. Long-range couplings from the hydroxyl proton OH-8 (δ 9.41) to the sp2 carbons C-7 (δ 116.3) and C-9 (δ 116.8) and the sp2 oxygenated carbon C-8 (δ 148.7) and from the methyl proton CH3-9 (δ 2.15) to C-8, C-9 and the sp2 oxygenated carbon C-9a (δ 142.4) were observed. Further long-range couplings from the sp2 methine proton H-2′′ (δ 5.38) to the methyl carbon C-4′′ (δ 17.3), from the methyl proton H3-3′′ (δ 1.76) to the sp2 carbon C-1′′ (δ 129.8) and from the methyl proton H3-4′′ (δ 1.84) to C-1′′ and the sp2 methine carbon C-2′′ (δ 127.0) suggested the presence of an isobutyl group containing the partial structure II. This isobutyl group was connected to the quaternary carbon C-6 (δ 134.0) by the observation of 3J cross peaks from H-2′′ and H3-4′′ to C-6 in HMBC experiments. A comparison of the NMR data between 1 and 2 revealed that the aromatic 13C chemical sift values were very similar, whereas the sp2 methine carbon at C-7 in 2 was replaced by a sp2 fully substituted carbon in 1, indicating that ring B in 1 had a hexasubstituted benzene ring and the aromatic proton at C-7 of 2 was substituted by a chlorine atom in 1 (Figure 2). (3) The molecular formula and degrees of unsaturation of 1 indicated that the aromatic rings A and B were connected via an ether linkage between C-4a and C-5a and an ester bridge between C-9a and C-11, suggesting a depsidone skeleton with a central seven-membered ring C. Taken together, the structure of 1 was elucidated as shown in Figure 1, that fulfilled the molecular formula and degrees of unsaturation.
Inhibition of SOAT isozymes using SOAT1- and SOAT2-CHO cells
Cell-based assay
The effects of 1 to 6 on cholesteryl ester (CE) synthesis were evaluated in a cell-based assay using SOAT1- and SOAT2-CHO cells. As shown in Table 3, 1 to 4 inhibited SOAT1 and SOAT2 isozymes with analogous IC50 values (2.0–16 μM), giving selectivity index (log (IC50 for SOAT1)/(IC50 for SOAT2)) values (−0.15 to 0.88) ranging between −1.00 and +1.00 (dual-type inhibition).12 Both 5 and 6 also showed very weak inhibition (40–50% inhibition at 23–25 μM) in both SOAT1- and SOAT2-CHO cells.
Enzyme assay
The inhibition of SOAT by 1 to 6 was evaluated in the enzyme assay using microsomes prepared from SOAT1- and SOAT2-CHO cells. As shown in Table 3, the IC50 and selectivity index values of 1 to 6 in the enzyme assay were consistent with those in the cell-based assay. Depsidones with no or one chloride in their molecules (1 to 4) exhibited dual-type SOAT inhibition, whereas those with two chlorides (5 and 6) had decreased inhibitory activity.
Inhibition of CE synthesis in mouse peritoneal macrophages
The effects of 1 to 6 on CE synthesis (synthesized by SOAT1) were investigated in mouse peritoneal macrophages according to our established method.19 CE accumulation in macrophages promotes atherosclerosis in arterial walls.20 Compounds 1 to 4 inhibited CE synthesis with IC50 values of 6.8, 12 and 19 μM, respectively (Table 3), consistent with those in SOAT1-CHO cells.
Antimicrobial activity
The antimicrobial activities of 1 to 6 (10 μg per disk), as determined by the agar diffusion assay, are summarized in Table 4. All compounds exhibited weak antimicrobial activity against the Gram-positive bacteria, Bacillus subtilis, Staphylococcus aureus and Micrococcus luteus, but did not affect the growth of the Gram-negative bacteria, Escherichia coli and Pseudomonas aeruginosa or yeast Candida albicans.
Conclusion
In the present study, a new chlorinated depsidone, 7-chlorofolipastatin (1), was isolated together with five known depsidones, 2 to 6, from the culture broth of a marine-derived Aspergillus strain. Compounds 1 to 4 moderately inhibited SOAT1 and SOAT2 isozymes (moderate dual-type SOAT inhibition).
Methods
General
Various NMR spectra were obtained using 400 MHz spectrometer (Agilent Technologies, Santa Clara, CA, USA). FAB-MS spectra were recorded on a JMS-700 Mstation (JEOL, Tokyo, Japan). Optical rotations were measured with a digital polarimeter (DIP-1000; JASCO, Tokyo, Japan). UV spectra were recorded on an 8453 UV-visible spectrophotometer (Agilent Technologies). IR spectra were recorded on a Fourier transform an FT-710 spectrometer (Horiba, Kyoto, Japan).
Materials
The [1-14C]Oleic acid (1.85 GBq mmol−1) was purchased from PerkinElmer (Waltham, MA, USA). Fetal bovine serum was purchased from Biowest (Nuaille, France). Dulbecco’s modified Eagle’s medium and Hank’s buffered salt solution were purchased from Nissui Pharmaceutical (Tokyo, Japan). GIT medium was from Nippon Seiyaku (Tokyo, Japan). Penicillin (10 000 units ml−1), streptomycin (10 000 mg ml−1) and glutamine (200 mm) solution were from Invitrogen (Carlsbad, CA, USA). Phosphatidylcholine, phosphatidylserine, dicetylphosphate, cholesterol, Ham’s F-12 medium, malachite green oxalate and BFA1 were purchased from Sigma-Aldrich (St Louis, MO, USA). Perchloric acid and Triton X-100 were purchased from Wako (Osaka, Japan).
Fungal strain and identification
The fungal strain NKH-007 was isolated from soil that was collected from the Suruga Bay, Japan (138°18.1207'E, 34°22.4813'N) at a depth of 331 m on 15 July 2013. In a BLAST search from the NCBI (National Center for Biotechnology Information), NKH-007 had 100% similarity with the rDNA sequence of the internal transcribed spacer region of Aspergillus ungui SRRC 344 (GenBank accession number AY373872).21 Therefore, the strain NKH-007 was identified with Aspergillus ungui.
Fermentation of Aspergillus ungui NKH-007
A loopful of spores of strain NKH-007 was inoculated into 100 ml seed medium consisting of 2.4% potato dextrose broth (Becton Dickinson and Company, Franklin Lakes, NJ, USA) and 0.1% agar (adjusted to pH 6.0 before sterilization) in a 500-ml Erlenmeyer flask. The inoculated Erlenmeyer flask was incubated in a rotary shaker (200 r.p.m.) at 27 °C for 5 days to obtain the seed culture. In order to produce 1, the culture was initiated by transferring 1 ml seed culture into each of six 500-ml culture bottles (AS ONE, Osaka, Japan) containing 100 ml production medium (2.4% potato dextrose broth, adjusted to pH 6.0 before sterilization). Fermentation was carried out under static conditions at 27 °C for 14 days.
Structural determination of 2 to 6
Using spectral data including 1H NMR, 13C NMR and MS, and the search results of SciFinder Scholar, 2 to 6 were identified as the known depsidones folipastatin (2), unguinol (3), 2-chlorounginol (4), 2,7-dichlorounginol (5) and nornidulin (6), respectively (Figure 1).
Cell culture
Two cell lines, CHO cells expressing SOAT1 and SOAT2 isozymes of African green monkey (SOAT1- and SOAT2-CHO cells, respectively),13 were kind gifts from Dr LL Rudel (Wake Forest University, Winston-Salem, NC, USA). Cells were maintained as described previously.12
Assays for SOAT activity in SOAT1- and SOAT2-CHO cells
Assays for SOAT1 and SOAT2 activities using SOAT1- and SOAT2-CHO cells were carried out by our established method.12 Briefly, SOAT1- or SOAT2-CHO cells (1.25 × 105 cells in 250 μl of medium) were cultured in a 48-well plastic microplate in the culture medium described above and allowed to recover overnight at 37 °C in 5% CO2. The assays were conducted with cells that were at least 80% confluent. Following overnight recovery, the test sample (2.5 μl MeOH solution) and [1-14C]oleic acid (5 μl 10% EtOH/phosphate-buffered saline solution, 1 nmol, 1.85 KBq) were added to each culture. The medium was removed after a 6-h incubation at 37 °C in 5% CO2, and the cells in each well were washed twice with phosphate-buffered saline. The cells were lysed by adding 0.25 ml of 10 mM Tris-HCl (pH 7.5) containing 0.1% (w/v) sodium dodecyl sulfate, and [14C]CE was analyzed with a BAS2000 analyzer (Fuji Film, Tokyo, Japan). In this cell-based assay, [14C]CE was produced by the reaction of SOAT1 or SOAT2. SOAT inhibitory activity (%) was defined as ([1-14C]CE-drug/[14C]CE-control) × 100. The IC50 value was defined as the drug concentration that inhibited biological activity by 50%.
Assay for SOAT activity in microsomes from SOAT1- and SOAT2-CHO cells
SOAT1 and SOAT2 activities were determined using microsomes prepared from SOAT1- and SOAT2-CHO cells as the enzyme source.12 Briefly, an assay mixture containing 2.5 mg ml−1 bovine serum albumin in buffer A and [1-14C]oleoyl-CoA (20 μM, 3.7 kBq) together with a test sample (added as a 10 μl methanol solution), and the SOAT1 or SOAT2 microsomal fraction (150 or 10 μg of protein, respectively) in a total volume of 200 μl were incubated at 37 °C for 5 min. The reaction was started by adding [1-14C]oleoyl-CoA and stopped by adding 1.2 ml of CHCl3/MeOH (2:1). The product [14C]CE was extracted by the method of Bligh and Dyer.22 After the organic solvent was removed by evaporation, lipids were separated on a TLC plate and the radioactivity of [14C]CE was measured as described above.
Assay for CE synthesis in mouse peritoneal macrophages
The assay for CE synthesis from [14C]oleic acid was carried out according to a previously described method.19 Briefly, mouse peritoneal macrophages (5.0 × 105 cells per 0.25 ml of medium B (containing Dulbecco’s modified Eagle’s medium supplemented with 8.0% (v/v) lipoprotein-deficient serum, penicillin (100 units ml−1) and streptomycin (100 mg ml−1)) were cultured in each well of a 48-well plastic microplate with a test compound (in 2.5 μl of CH3CN) and liposomes (10.0 μl, 1.0 μmol phosphatidylcholine, 1.0 μmol phosphatidylserine, 0.20 μmol dicetylphosphate and 1.5 μmol cholesterol, suspended in 1.0 ml of 0.30 M glucose) together with [14C]oleic acid (5.0 μl (1.85 kBq) in 10% ethanol/phosphate-buffered saline solution). Following a 14-h incubation, cellular lipids were extracted to measure the radioactivity of [14C]CE according to the same method as described above.
Antimicrobial activity
The antimicrobial activity of a sample against six species of microorganism was measured by the agar diffusion method using paper disks. Media for microorganisms were as follows: nutrient agar (Sanko Junyaku, Tokyo, Japan) for B. subtilis PCI219, S. aureus FDA209P, M. luteus KB212, E. coli JM109 and P. aeruginosa IFO12689, and medium composed of 1.0% glucose, 0.50% yeast extract and 0.80% agar for C. albicans ATCC90029. A paper disk (i.d. 6 mm; Toyo Roshi Kaisha, Tokyo, Japan) containing a sample (10 μg) was placed on the agar plate. Bacteria were incubated at 37 °C for 24 h. C. albicans was incubated at 27 °C for 48 h. Antimicrobial activity was expressed as the diameter (mm) of the inhibitory zone.
Accession codes
References
Rudel, L. L., Lee, R. G. & Cockman, T. L. Acyl coenzyme A: cholesterol acyltransferase types 1 and 2: structure and function in atherosclerosis. Curr. Opin. Lipidol. 12, 121–127 (2001).
Alegret, M., Llaverias, G. & Silvestre, J. S. Acyl coenzyme A:cholesterol acyltransferase inhibitors as hypolipidemic and antiatherosclerotic drugs. Methods Find. Exp. Clin. Pharmacol. 26, 563–586 (2004).
Tardif, J. C. et al. Effects of the acyl coenzyme A:cholesterol acyltransferase inhibitor avasimibe on human atherosclerotic lesions. Circulation 110, 3372–3377 (2004).
Nissen, S. E. et al. Effect of ACAT inhibition on the progression of coronary atherosclerosis. N. Engl. J. Med. 354, 1253–1263 (2006).
Meuwese, M. C. et al. ACAT inhibition and progression of carotid atherosclerosis in patients with familial hypercholesterolemia: the CAPTIVATE randomized trial. JAMA 301, 1131–1139 (2009).
Chang, C. C., Huh, H. Y., Cadigan, K. M. & Chang, T. Y. Molecular cloning and functional expression of human acyl-coenzyme A:cholesterol acyltransferase cDNA in mutant Chinese hamster ovary cells. J. Biol. Chem. 268, 20747–20755 (1993).
Anderson, R. A. et al. Identification of a form of acyl-CoA:cholesterol acyltransferase specific to liver and intestine in nonhuman primates. J. Biol. Chem. 273, 26747–26754 (1998).
Cases, S. et al. ACAT-2, a second mammalian acyl-CoA:cholesterol acyltransferase. Its cloning, expression, and characterization. J. Biol. Chem. 273, 26755–26764 (1998).
Oelkers, P., Behari, A., Cromley, D., Billheimer, J. T. & Sturley, S. L. Characterization of two human genes encoding acyl coenzyme A:cholesterol acyltransferase-related enzymes. J. Biol. Chem. 273, 26765–26771 (1998).
Parini, P. et al. ACAT2 is localized to hepatocytes and is the major cholesterol-esterifying enzyme in human liver. Circulation 110, 2017–2023 (2004).
Tomoda, H. & Ōmura, S Potential therapeutics for obesity and atherosclerosis: inhibitors of neutral lipid metabolism from microorganisms. Pharmacol. Ther. 115, 375–389 (2007).
Ohshiro, T., Rudel, L. L., Ōmura, S . & Tomoda, H Selectivity of microbial acyl-CoA:cholesterol acyltransferase inhibitors toward isozymes. J. Antibiot. 60, 43–51 (2007).
Lada, A. T. et al. Identification of ACAT1- and ACAT2-specific inhibitors using a novel, cell-based fluorescence assay: individual ACAT uniqueness. J. Lipid Res. 45, 378–386 (2004).
Hamano, K. et al. Folipastatin, a new depsidone compound from Aspergillus unguis as an inhibitor of phospholipase A2. Taxonomy, fermentation, isolation, structure determination and biological properties. J. Antibiot. 45, 1195–1201 (1992).
Feighner, S. D., Salituro, G. M., Smith, J. L. & Tsou, N. N. Unguinol and analogs are animal growth permittants. US patent 5,350,763 (1994).
Kawahara, N., Nakajima, S., Satoh, Y., Yamazaki, M. & Kawai, K. Studies on fungal products. XVIII.: isolation and structures of a new fungal depsidone related to nidulin and a new phthalide from Emericella unguis. Chem. Pharm. Bull. 36, 1970–1975 (1988).
Sureram, S. et al. Depsidones, aromatase inhibitors and radical scavenging agents from the marine-derived fungus Aspergillus unguis CRI282-03. Planta. Med. 78, 582–588 (2012).
Dean, F. M., Roberts, J. C. & Robertson, A. Chemistry of fungi. XXII. Nidulin and nornidulin (ustin): chlorine-containing metabolic products of Aspergillus nidulans. J. Chem. Soc. (1954) 1432–1439 (1954).
Namatame, I., Tomoda, H., Arai, H., Inoue, K. & Ōmura, S Complete inhibition of mouse macrophage-derived foam cell formation by triacsin C. J. Biochem. 125, 319–327 (1999).
Ouimet, M. & Marcel, Y. L. Regulation of lipid droplet cholesterol efflux from macrophage foam cells. Arterioscler. Thromb. Vasc. Biol. 32, 575–581 (2012).
Haugland, R. A., Varma, M., Wymer, L. J. & Vesper, S. J Quantitative PCR analysis of selected Aspergillus Penicillium and Paecilomyces species. Syst. Appl. Microbiol. 27, 198–210 (2004).
Bligh, E. G. & Dyer, W. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Acknowledgements
We thank Ms N Sato and Dr K Nagai, School of Pharmacy, Kitasato University, for measurements of mass and NMR spectra. This work was supported by JSPS KAKENHI Grant Number 26253009 (to HT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Uchida, R., Nakajyo, K., Kobayashi, K. et al. 7-Chlorofolipastatin, an inhibitor of sterol O-acyltransferase, produced by marine-derived Aspergillus ungui NKH-007. J Antibiot 69, 647–651 (2016). https://doi.org/10.1038/ja.2016.27
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2016.27
This article is cited by
-
Haneummycin, a new 22-membered macrolide lactam antibiotic, produced by marine-derived Streptomyces sp. KM77-8
The Journal of Antibiotics (2023)
-
A Mixture of Atropisomers Enhances Neutral Lipid Degradation in Mammalian Cells with Autophagy Induction
Scientific Reports (2018)
-
Aspergone, a new chromanone derivative from fungus Aspergillus sp. SCSIO41002 derived of mangrove soil sample
The Journal of Antibiotics (2017)