Abstract
The major histocompatibility complex (MHC) plays a central role in the adaptive immune response and is the most polymorphic gene family in vertebrates. Although high-throughput sequencing has increasingly been used for genotyping families of co-amplifying MHC genes, its potential to facilitate early steps in the characterisation of MHC variation in nonmodel organism has not been fully explored. In this study we evaluated the usefulness of de novo transcriptome assembly in characterisation of MHC sequence diversity. We found that although de novo transcriptome assembly of MHC I genes does not reconstruct sequences of individual alleles, it does allow the identification of conserved regions for PCR primer design. Using the newly designed primers, we characterised MHC I sequences in the bank vole. Phylogenetic analysis of the partial MHC I coding sequence (2–4 exons) of the bank vole revealed a lack of orthology to MHC I of other Cricetidae, consistent with the high gene turnover of this region. The diversity of expressed alleles was characterised using ultra-deep sequencing of the third exon that codes for the peptide-binding region of the MHC molecule. High allelic diversity was demonstrated, with 72 alleles found in 29 individuals. Interindividual variation in the number of expressed loci was found, with the number of alleles per individual ranging from 5 to 14. Strong signatures of positive selection were found for 8 amino acid sites, most of which are inferred to bind antigens in human MHC, indicating conservation of structure despite rapid sequence evolution.
Similar content being viewed by others
Introduction
The major histocompatibility complex (MHC) is a gene family that encodes a group of transmembrane proteins. The highly polymorphic classical MHC class I (Ia) and II genes play a major role in pathogen recognition, whereas a number of nonclassical MHC class I-like (Ib), class II-like and class III genes (of limited polymorphism) encode several other immune-related proteins. The classical genes of class I and II differ in structure, antigen-processing pathway and tissue distribution. MHC class I genes are expressed in every nucleated cell and present antigens derived from intracellular pathogens, whereas MHC class II are mainly expressed by specialised antigen-presenting cells and binds peptides of exogenous origin (Klein, 1986).
The classical MHC genes are the most polymorphic genes of jawed vertebrates. This high degree of polymorphism is thought to result from selective pressure on hosts exerted by parasites (Apanius et al., 1997; Yeager and Hughes, 1999; Jeffery and Bangham, 2000). Indeed, theoretical work showed that such selection is best explained by the evolutionary arms race between hosts and parasites, whereby novel MHC alleles confer resistance to fast-evolving parasites (Ejsmond and Radwan, 2015). The majority of the polymorphic sites in MHC proteins are located in codons that form the antigen-binding site (Reche and Reinherz, 2003) where an excess of nonsynonymous versus synonymous substitutions (dN/dS) is the signature of positive selection.
The number of expressed MHC genes differs greatly between species, but the reasons for this variation are unclear. In contrast to the limited number of loci found in some vertebrates, including model organisms like human, mouse and chicken (reviewed by Kelley et al., 2005), some other groups, for example, passerine birds (Westerdahl et al., 2005; Zagalska-Neubauer et al., 2010) and several neoteleost fishes (Miller et al., 2002), express many MHC loci. Furthermore, in model hominoids and rodents, MHC class I genes evolve faster than those in class II, resulting in a rapid loss of orthology between class I genes across different orders (Yeager and Hughes, 1999), and even within the same order (Hughes and Nei, 1989a). Lack of such orthologous relationships has been explained by nonmutually exclusive processes, such as concerted evolution homogenising alleles at the species level (Rada et al., 1990) or faster rate of duplication (Hughes and Nei, 1989a). However, such trends are not always evident or reflected in the higher dN/dS ratios that would indicate positive selection. For example, in sciurids and ursids, positive selection on MHC class I genes appears to be weaker than on MHC class II, suggesting the former are under relaxed evolutionary pressure (Kuduk et al., 2012a, b).
Generalisations concerning the evolution of MHC class I genes are hindered by the scarcity of data on the number of expressed genes, their polymorphism and their molecular evolution. The reason is twofold. First, conserved regions suitable for the design of PCR primers are often shared between loci, meaning that alleles from many loci may co-amplify (Babik, 2010). The challenge of genotyping such complex systems has only recently been overcome through the application of high-throughput sequencing (HTS) for MHC typing (Babik et al., 2009; Radwan et al., 2012), and new genotyping strategies are constantly being developed and improved (Lighten et al., 2014; Sebastian et al., 2016). The second reason is that projects involving nonmodel species are impeded by a lack of initial information about the system of interest. This problem is particularly serious for MHC class I loci, whose rapid evolution often leads to a loss of orthology to genes of related species. For instance, in members of the same rodent subfamily (Murinae), no orthology was found between classical (Ia) MHC genes in mice and rats (H2-K/D/L loci and RT1-A/CE loci, respectively; Hurt et al., 2004). Consequently, information from related species might not provide a useful starting point for characterisation of MHC class I sequences. Here, we overcome this difficulty through a primer design strategy based on de novo transcriptome assembly. We chose transcriptome rather than a draft genome (recently available for bank vole, GCA_001305785.1, and few other Arvicolinae species) for two main reasons. First, the MHC region is known to possess a high number of pseudogenes that cannot be distinguished from expressed MHC genes without a transcriptome analysis. Second, genome assemblies of nonmodel species are usually highly fragmented and error prone, especially in highly duplicated, variable regions, such as MHC. Even in the best-studied system (humans), cloning and Sanger sequencing (as opposed to HTS) was used to characterise MHC in the 1000 Genomes Project (Gourraud et al., 2014). In the case of nonmodel species, de novo assembled transcriptomes have already been used in the identification of genetic markers for population genetics and phylogenetics (for example, in newts; Zieliński et al., 2014), but their utility in primer design for multigene families has not yet been explored.
We used this new strategy to design primers for MHC class I genes of the bank vole, Myodes glareolus (also known as Clethrionomys glareolus). The bank vole is a small rodent common in Europe and northern Asia, and widely used in ecological and evolutionary surveys (see, for example, Mokkonen et al., 2011; Filipi et al., 2015; Konczal et al., 2016), including studies of adaptive evolution in gene families (for example, haemoglobins, Marková et al., 2014; Kotlík et al., 2014). Unlike MHC class II, in which polymorphism, the molecular signatures of selection and its associations with parasites have been extensively studied (Bryja et al., 2006; Axtner and Sommer, 2007; Babik and Radwan, 2007; Kloch et al., 2010; Scherman et al., 2014), we lack similar data on MHC class I. This is unfortunate, as (among other reasons) the bank vole is a reservoir species for Puumala virus, one of the pathogenic hantaviruses, that causes in humans a mild form of haemorrhagic fever with renal syndrome (Vaheri et al., 2013). Several components of the bank vole’s immune system have been analysed in the context of Puumala virus, such as the association with MHC class II alleles (Deter et al., 2008) and the expression of tumour necrosis factor-α (Guivier et al., 2010). Considering the important role that MHC class I plays in the presentation of antigens derived from intracellular pathogens (like viruses), correlation with MHC class I is highly desirable, yet missing because of the lack of genetic resources.
The first aim of our study was to establish transcriptome assembly as a tool for the initial characterisation of polymorphic genes (such as MHC) in nonmodel species. Because de novo assembly of highly polymorphic gene families is notoriously difficult, we first benchmarked de novo MHC class I transcriptome predictions using previously published RNA sequencing (RNA-Seq) data from mice (Grabherr et al., 2011; Hutchins et al., 2012), the best-studied rodent model, with a fully sequenced MHC region. The predicted transcripts were compared with mouse allele reference sequences of MHC class I loci (K and D). Subsequently, we reconstructed de novo transcriptomes for seven bank vole individuals and designed primers in conserved regions of the MHC class I transcripts. We designed two sets of primers, one that would amplify almost the entire coding sequence of MHC class I transcript, and the second for the third exon only.
Our second aim was to provide a comprehensive characterisation of the MHC class I genes in the bank vole. We examined the orthology with other rodents using fragments of long transcripts that encode the extracellular part of the MHC molecule (2–4 exons). Furthermore, using the third exon of MHC class I, which codes for a part of the peptide-binding region, we described the variation in the number of expressed alleles, sequence diversity and signatures of positive selection.
Materials and methods
Validation of de novo MHC class I transcriptome predictions in the house mouse
To evaluate the potential of de novo transcriptome assembly of duplicated and polymorphic genes, such as MHC class I, we selected two independent RNA-Seq data sets from the house mouse. The first murine RNA-Seq consisted of 105.3 million 75 bp paired-end reads sequenced with an Illumina Genome Analyzer II (San Diego, CA, USA) from unstimulated dendritic cells of the strain C57BL/6J (NCBI Bioproject PRJNA140057, run SRR203276) (Grabherr et al., 2011). The second RNA-seq contained 13.8 million 90 bp paired-end reads obtained with an Illumina HiSeq 2000 from anti-CD3/anti-CD28 treated CD4+ T cells (4 h) of the mouse strain C57BL/6JJcl (NCBI Bioproject PRJNA151367, run SRR391077) (Hutchins et al., 2012).
The Trinity assembler (Grabherr et al., 2011, version 2013-10-11, default parameters) was chosen for de novo assembly of the two murine RNA-seq experiments because independent evaluations of de novo assemblers ranked Trinity on the top (Zhao et al., 2011; Li et al., 2014). We note that the first mouse data set was assembled by Grabherr et al. (2011) in the original publication of the Trinity tool, with outstanding results. In that study, Trinity fully reconstructed a large portion of transcripts, including alternatively spliced isoforms and transcripts from recently duplicated genes. Specifically, Trinity matched the full-length transcripts for 463 (61.6%) of 752 paralogous transcripts in the control set (>70% identity between paralogues). However, the quality of assembly of MHC genes has not yet been reported.
Three assembly strategies were tested in the present study: (1) direct assembly of the full set of RNA-Seq reads, (2) assembly of the subset of reads that align with mouse MHC class I reference sequences and (3) assembly of a subset of reads that had previously been mapped to human MHC class I reference sequences. The second and third strategies were carried out to test whether assembly quality can be improved by the reduction of complexity of the reads. For this purpose, we retrieved a subset of targeted reads based on similarity to transcripts from (1) the mouse itself and (2) humans. In the third strategy, we used a relatively distant mammalian species (from a different order) to see how the filtering performs if no data from close relatives are available. To retrieve targeted mouse reads, all reads were mapped using the Bowtie2 tool v2.2.0 (Langmead and Salzberg, 2012) in local alignment mode to the reference sequences. Murine references consisted of full MHC complementary DNAs (cDNAs) extracted from Genbank (Benson et al., 2013) corresponding to the known alleles of the laboratory mouse strains analysed (H2-K1: NM_001001892.2, H2-D1: NM_010380.3). Human references consisted of 7996 MHC class I alleles extracted from the IMGT/HLA database (Robinson et al., 2013) (access: April 2014). In addition, three control genes without paralogues were added to the reference set and included in further analyses: β-2 microglobulin (B2M: NM_009735.3) and the two isoforms of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH-iso1: NM_001289726.1 and GAPDH-iso2: NM_008084.3).
The contigs retrieved by each assembly strategy were aligned to the known MHC class I alleles of the laboratory mouse, H2-K1 (NM001001892.2) and H2-D1 (NM010380.3), using BLASTN (Altschul et al., 1990). Contigs matching for >75% of a reference sequence length with an identity of >80% were considered to be recovered transcripts and used for further analysis. The percentage of reference sequence length covered by a recovered transcript was calculated as the number of identical nucleotides in the best alignment divided by the length of the reference sequence matched.
De novo assembly of bank vole transcriptomes and evaluation of MHC I sequence predictions
Because de novo transcriptome assembly (first strategy) fitted predictions for MHC transcripts in mouse better than strategies based on prefiltering (see Results), we applied the first strategy for bank vole MHC class I reconstruction. Direct de novo assemblies of bank vole spleen transcriptomes were performed using sequences from seven voles collected in Great Britain (Aberdeenshire and Gloucestershire) in 2009 and 2011. Voles were trapped and taken under Natural England (general) and Scottish Natural Heritage (10080) licences.
Spleen samples were stored in RNAlater (QIAGEN, Hilden, Germany), and total RNA was extracted with the RNeasy Mini Kit (QIAGEN). This was followed by DNase treatment using the TURBO DNA-free Kit (Ambion, Austin, TX, USA) and an additional clean-up step using the RNeasy Mini Kit. Library preparation and sequencing were performed with the use of the standard Illumina protocols on the Illumina HiSeq 2000 as described previously (Marková et al., 2014, 2015). Briefly, after poly(A) enrichment and fragmentation, the RNA was size selected to 250–400 bp, reverse transcribed into cDNA, end repaired and PCR enriched. The resulting libraries were sequenced using the 100 bp paired-end module.
We performed direct assembly of RNA-Seq reads for each of the seven bank vole individuals with the Trinity assembler (version 2013-10-11, Grabherr et al., 2011). The assembled contigs matching MHC class I were recovered with BLASTN 2.2.29 (Altschul et al., 1990) to the mouse reference sequences H2-K1 (1669 bp, NM001001892.2) and H2-D1 (1736 bp, NM010380.3). The retrieved sequences were later used for primer design. After successful amplification of cDNA with newly developed primers, we retrospectively evaluated the bank vole transcriptomic MHC class I predictions as explained above for the mouse, but using the cloned and Sanger-sequenced cDNA sequences from three bank vole individuals from set ‘A’ (see ‘Amplification and cloning of MHC I transcripts of the bank vole’) as a reference. Contigs with a minimum identity of 80% over at least 75% of the length of a reference sequence were considered (and referred to hereafter) as MHC recovered transcripts.
Primer design for MHC class I loci in the bank vole
Multiple alignment (MAFFT; Katoh, 2002) of the bank vole recovered transcripts with the murine references was used to design primers in highly conserved regions, with priority given to the lack of polymorphism in transcriptome contigs (Supplementary Figure S2). Exon boundaries were delineated based on homology with a mouse H2-K1 reference sequence (NCBI refseq: NM_001001892.2). As the last step, the chosen primer region was visualised by mapping reads on the assemblies (Tablet software, version 1.13.12.17; Milne et al., 2013) to ensure it was free of single-nucleotide polymorphisms that could have been obscured by the assembly process. All primers were designed with the assistance of FastPCR software (Kalendar et al., 2014) in order to optimise oligonucleotide properties and avoid primer-dimer formation. In all experiments primers were used to amplify cDNA.
The first primer pair (MyglMHCI_F, MyglMHCI_R) amplified a majority of the coding sequence of the MHC class I transcript (Figure 1). The forward primer, MyglMHCI_F (5′-GSTGCTCCTRCTGCTGGC-3′), was located in the first exon that encodes the leading peptide. The reverse primer, MyglMHCI_R (5′-CTGGATGTCACAGGAGAGACC-3′), was located in the 3′ untranslated region. The expected length of the product exceeds 1100 bp, and was not constant. This variability may result from length differences in exons 5–8 among loci (Crew et al., 1991). In addition, this primer pair may occasionally co-amplify nonclassical loci with shortened cytosolic tails (Hughes and Nei, 1989a), but those sequences should be easy to identify, both by using BLAST searches and by considering that nonclassical genes are much less polymorphic than classical ones. These primers were used to obtain long transcripts for comparison with de novo assembled contigs and for the phylogenetic analysis.
Location of PCR primer pairs. Positions of forward and reverse primers are shown on the schematic diagram of the class I MHC mRNA transcript. Each exon corresponds to a distinct domain of the MHC molecule. The first exon encodes the leader peptide that is cleaved off post-translationally. Exons 2 and 3 encode the α1 and α2 domains, respectively, that are the most polymorphic regions and form the antigen-binding groove. Exon 4 encodes the α3 domain, a conserved region of the MHC I molecule. Exons 5 to 8 encode the transmembrane domain and cytosolic tail.
The second pair of primers (MyglMHCI3_F, MyglMHCI3_R) was developed to characterise the third exon of the MHC class I gene, as it encodes one of the antigen-binding groove domains (α2). To avoid co-amplification of genomic DNA during cDNA amplification, the primers were located on exon borders or in the adjacent exons (Figure 1). The initially designed primers were further checked against cloned transcripts (see below). The forward (MyglMHCI3_F: 5′-ACTACAACCAGAGCGAGG-3′) and reverse primers (MyglMHCI3_R: 5′-TGTGCCTTTGGGSGAWCT-3′) amplified a 277 bp-long fragment of the MHC class I transcript (complete third exon) and these products were used for polymorphism and selection analysis.
Amplification and cloning of MHC I transcripts of the bank vole
To characterise the MHC class I transcripts, we used cDNA from three of the seven voles used for the transcriptome assembly (‘set A’), as well as three vole samples obtained from a laboratory colony (‘set B’; courtesy P Koteja, Krakow, Poland) that was derived from a wild vole population in the Niepolomice Forest, Poland (Sadowska et al., 2008). The breeding and experimental protocols were approved by the Polish State and Local Ethical Committee for Ethics in Animal Research in Krakow (decision nos. 99/2006, 21/2010 and 22/2010). We added individuals from the laboratory colony to ensure that our primers would capture MHC alleles in individuals from different populations. Livers and spleens from the voles were obtained during necropsy and preserved in RNAlater (Sigma-Aldrich, St Louis, MO, USA). Total RNA was extracted using RNAzol RT (Sigma-Aldrich) according to the manufacturer’s instructions. A FastPrep-24 Instrument (Bio Lysing Matrix D Tubes; MP Biomedicals, Strasbourg, France) was used for homogenisation of tissues during RNA isolation. Extracted RNA was reverse transcribed with oligo(dT) primers and the Maxima H Minus First-Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). Up to 5 μg of total RNA was used for transcription.
For amplification of MHC class I messenger transcripts, cDNA HotStarTaq Master Mix (QIAGEN) was used together with 0.2 μM of each primer (MyglMHCI_F, MyglMHCI_R) and 2 μl of transcribed cDNA. The reaction volume was 15 μl. The PCR was conducted using the following conditions: initial denaturation at 95 °C (15 min); 30 cycles of denaturation at 95 °C (20 s), annealing at 53 °C (35 s) and extension at 72 °C (1 min and 20 s); and final extension at 72 °C (10 min). The PCR products were separated on 1.5% agarose gel and bands of the expected length (∼1100–1200 bp) were excised using the QIAquick Gel Extraction Kit (QIAGEN) according to the manufacturer’s instructions. Inserts were cloned with the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The amplification and cloning procedure was performed twice for each sample.
For the ‘set A’, 50 colonies per individual (sum of two cloning procedures, 20–30 colonies for each) were picked based on a blue-white screening test. When positive transformation was confirmed by colony PCR (FIREPol 5x Master Mix, Solis Biodyne, Tartu, Estonia) with the M13 vector primers), 82 clones (25–29 per individual) were sequenced in both directions by Sanger sequencing. M13 forward and reverse primers were used and the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on a 3130xl Genetic Analyzer (Applied Biosystems). For ‘set B’, altogether between 18 and 40 colonies were picked in two cloning procedures, and 57 clones (13–31 per individual) were sequenced as described above. Cloned sequences were manually curated (Chromas Lite 2.1.1, Technelysium, Brisbane, QLD, Australia) and merged (MERGER, EMBOSS; Rice et al., 2000).
We accepted a sequence as a true variant if it was present in two independent PCRs (from the same or different individuals). In addition, to retrieve more transcripts, we used CONS with default parameters (EMBOSS; Rice et al., 2000) to create consensus variants that represented highly similar (1 bp difference), but not identical, clones. Between 3 and 14 sequences were used to create each consensus variant. The remaining unique clones were discarded as possible chimeras. All sequences that were accepted as true variants are listed in Supplementary File S1.
MHC class I phylogenetic analysis
Sequences of cloned full-length transcripts of the bank vole MHC class I (subfamily Arvicolinae), along with available sequences from the Cricetidae family were used for phylogenetic analysis: subfamily Cricetinae—Cricetulus griseus, Chinese hamster: NM_001243995, NM_001243994.1, NM_001243993.1, NM_001243997.1; subfamily Neotominae—Peromyscus maniculatus, North American deer mouse: U12887.1, U12886.1, U12885.1, U16846.1, U16847.1, U12822.3; subfamily Sigmodontinae—Sigmodon hispidus, hispid cotton rat: AF480859.1) and Muridae family (Mus musculus, house mouse H2-K1 and H2-L reference sequences: NM_001001892.2, NM_001267808.1). Human MHC (human leukocyte antigen (HLA)) was used as an outgroup (HLA-A, gi: 337752171; HLA-B, gi: 170650640; HLA-C, gi: 339882742). As exons 1 and 5–8 are not easily aligned, and the region spanned by the deposited sequences was variable, only fragments of exons 2–4 were used in this analysis. A maximum likelihood tree was constructed in MEGA 6.0 (Tamura et al., 2013), assuming the GTR+I+G model and with 5000 bootstrap replicates.
Bank vole MHC class I exon 3 sequencing
For a more comprehensive characterisation of MHC class I exon 3 sequences, capturing population-level diversity, 29 voles from a laboratory colony derived from the Niepolomice Forest population (the same as ‘set B’) were used. Spleens were obtained during necropsy; RNA was extracted and cDNA was synthesised as described above. Independent extraction and amplification was performed for two spleen fragments for each individual. Amplicons from the first and the second spleen fragment were sequenced as independent libraries, subsequently referred to as I1 and I2.
Amplicon libraries for HTS of the third exon were prepared using the fusion PCR method. Fusion forward primers contained Ion Torrent adaptor A (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′), a 6 bp tag and the forward target primer (MyglMHCI3_F). Fusion reverse primers contained Ion Torrent adaptor trP1 (5′-CCTCTCTATGGGCAGTCGGTGAT-3′), a 6 bp tag and the designed reverse target primer (MyglMHCI3_R). The tags were custom-made; the combination of two tags was unique for each individual and allowed us to demultiplex samples after sequencing.
PCR was performed in 10 μl reaction volumes with HotStar MasterMix (QIAGEN), 1.25 μM of degenerate forward primer, 0.75 μM of non-degenerate reverse primer and 1 μl of transcribed cDNA.
The cycling conditions were: 95 °C for 15 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 60 s and 72 °C for 25 s, and a final extension at 72 °C for 10 min. Products were pooled into approximately equimolar quantities, based on concentration estimation with agarose gel electrophoresis. To purify and ascertain the desired length of products, pools were separated on 1.5% agarose gel, and appropriate bands were excised using the QIAquick Gel Extraction Kit (QIAGEN). Templates for HTS were prepared with the Ion PGM Template OT2 200 Kit, emulsion PCR and enrichment on an Ion OneTouch 2 System (Life Technologies, Carlsbad, CA, USA), and sequencing on an Ion Torrent PGM apparatus (Life Technologies) with the Ion PGM Sequencing 400 Kit. Sequencing was performed either at the facilities of Adam Mickiewicz University, Poznan, Poland (314 v2 or 318™ v2 chips, I1 and part of I2) or in the Functional Genomics Center, Uni/ETH Zurich, Switzerland (318 v2 chip, part of I2).
Bank vole MHC class I exon 3 genotyping
Although several protocols are available for the genotyping of MHC in nonmodel species using data obtained by HTS (reviewed in Lighten et al., 2014), none of them has been tested on a large number of cDNA samples. Differences in expression levels may affect variant frequencies in amplicons that adds to the existing variability caused by varying amplification efficiencies (Sommer et al., 2013) or systematic sequencing errors (McElroy et al., 2012). As such variation may affect common genotyping assumptions (for example, the number of reads yielded by an artefact of a highly expressed allele may exceed the number of reads yielded by a low-expressed, real allele), we used a conservative modification of previously published protocols. First, we implemented artefact clustering to mitigate the high error rates of HTS, a method that was proven to be successful in the genotyping of complex, polymorphic systems (Sebastian et al., 2016). Second, final genotypes were inferred based on the comparison of sequencing results from two independent replicates, each from a separate isolation from one of two spleen fragments for each individual. In order to be accepted as a true allele, a variant had to be present in both replicates. We hereafter use the term ‘allele’ to describe all co-amplifying genetic variants from different, duplicated MHC class I loci in a given individual. Although this is not a strict use of the term (that is, referring to genetic variants present at a particular locus), such terminology is widely applied in MHC literature when allele assignment to loci is not possible (see, for example, Bryja et al., 2006; Babik, 2010). Moreover, as increasing the depth of sequencing also increases the chances of obtaining identical sequencing errors (especially in the case of systematic, platform-specific errors), we additionally required that a given variant occur in at least one of the replicates at a minimum per-amplicon frequency (PAF). This threshold was chosen to exclude variants that occurred in both replicates at PAFs lower than those of most variants that occurred in only one replicate (see Results, Figure 3). The rationale for this criterion is that although the majority of the erroneous sequences should be clustered with their parental allele, some may still be present after the clustering and filtering steps (for example, complex chimeras, contaminants or errors beyond clustering algorithm capacities).
Comparison of assigned variants’ PAFs (%) with an indication of whether they were present in both (I1_common, I2_common) or only one replicate (I1_unique, I2_unique). Assignment of variants to individuals was performed by AmpliSAS, after demultiplexing, clustering and filtering of chimeric sequences.
For final genotyping, we used the following clustering parameters: 0.6% substitution error rate (to allow up to 2 mismatches in a sequence of 277 bp), 1% indel error rate (for non-homopolymer indels, as homopolymer ones are clustered by default by AmpliSAS, Sebastian et al., 2016), only variants of correct or in-frame (discrete deviations of 3 bp from expected) length could be the dominant sequence of a cluster, minimum frequency with respect to the dominant: 25% (for detailed description of the parameters see Sebastian et al., 2016). To reduce computational load, the maximum number of clusters formed was limited to 60. Afterwards, we filtered chimeras using the filtering algorithm of AmpliSAS. These steps were conducted separately for the two data sets (I1 and I2), and the results were compared with the accompanying software of AmpliSAS AmpliCOMPARE (http://evobiolab.biol.amu.edu.pl/amplisat/).
Retrieved sequences were aligned with MAFFT 7 (Katoh, 2002; Katoh and Standley, 2013), and all but four agreed with the reading frame of a reference mouse H2-K1 and H2-D1 sequences. As the analysed data came from cDNA samples, the presence of a 1 bp deletion, or compensating deletion and insertion that would severely alter the reading frame, is highly unlikely. On the other hand, Ion Torrent technology is prone to high-frequency indels, that is, indels that are not random, occur at a given base position in the reference genome and more prevalent around particular sequence motifs (Bragg et al., 2013). Indeed, close examination of the clusters containing these sequences suggested they were most likely artefacts bearing indels that were artificially introduced during the alignment step of AmpliSAS clustering. As the corresponding correct sequences were present in other individuals, we manually corrected them.
Selection analysis in the exon 3 of bank vole MHC class I sequences
We analysed selection acting on MHC class I exon 3 in the bank vole using CODEML from the PAML4 package (Yang, 2007). CODEML identifies selection patterns through phylogenetic analyses of DNA sequences using the maximum likelihood method. Positive selection is inferred when ω>1, where ω=dN/dS (number of nonsynonymous over synonymous substitutions). We applied different statistical models of variable selective pressure among sites (see Supplementary File S2), which evaluated the ω ratio for each codon separately, as in most of the functional genes most residues are highly conserved and averaging ω over the sequence might not reveal a significant signature of positive selection. Likelihood ratio tests were performed, comparing nested models as follows: 2(Lb−La), where La is the likelihood of a model that does not allow for ω>1, and Lb is the most general one that does. The likelihood ratio tests were compared with the χ2 distribution with the degrees of freedom equal to the difference in the number of parameters between the two models. Posterior probabilities for site classes were calculated by the Bayes empirical Bayes method (Yang et al., 2005). In all models we used a phylogenetic tree generated in jModelTest2, with the best-fit model (GTR+G) identified by Akaike information criterion (Guindon and Gascuel, 2003; Darriba et al., 2012).
Results
Validation of de novo MHC class I transcriptome predictions in house mouse
The best transcriptome MHC class I predictions (that is, most similar to the mouse strains H2-K1 and H2-D1) were obtained by the direct Trinity assembly and by the assembly of filtered reads mapping to the mouse references; both strategies (the first and the second) performed with similar accuracy (Supplementary Table S1). The assembly of filtered reads that were aligned to human references (the third assembly strategy) produced a lower number of contigs that matched MHC alleles than the other two strategies; in addition, these contigs were shorter. This most likely resulted from the fact that the number of reads to assemble was drastically reduced in the case of filtering based on alignments to HLA. In the first murine RNA-Seq data set, originally containing 105.3 million reads, 601 000 and 88 000 matched the mouse and human references, respectively. In the smaller second murine RNA-Seq data set of 13.9 million reads, these numbers decreased to 73 000 and 18 000, respectively.
Statistics of the number of recovered transcripts and the percentages of reference sequence lengths covered are listed in Supplementary Table S1 and sequence alignments to the mouse MHC reference sequences are shown in Supplementary Figure S1. As shown in Supplementary Figure S1, the MHC transcripts most similar to the reference did not represent any specific locus; rather, they represented chimeras of H2-K1 and H2-D1 genes. However, the control sequences, B2M and GAPDH-iso2, perfectly matched a unique assembled transcript. Our benchmarking thus showed that direct de novo assembly of RNA-Seq data produces only partial reconstructions of the MHC class I family transcripts (also observed in MHC class II, data not shown). The predictions included chimeric reconstructions of highly homologous sequences from different loci. Nevertheless, they contained highly conserved regions that were identical to the references and could be used to design primers or probes to analyse the full set of gene members of the MHC family.
De novo assembly of bank vole transcriptomes and evaluation of MHC I sequence predictions
From several millions of paired-end reads for each individual and several thousands of transcript predictions (from 82 079 to 196 817), the number of contigs matching the mouse and rat MHC class I reference sequences with high identity (minimum identity of 80% covering at least 75% of the length) was quite low (see Table 1). Out of 7 individuals, in 3 we found 30 or more assembled contigs that resembled MHC class I references within the specified threshold; in another 2 individuals, only a few such contigs were present, and in the remaining 2 voles, no putative MHC class I contig was found (Table 1). The apparent lack of contigs for MHC class I in some individuals resulted from the quality threshold we set (see above). With relaxed criteria, fragments of MHC class I transcripts were present for all voles, for example, with minimal contig length of 500 bp, and with identity relaxed to 75% along half of the reference sequence, we retrieved between 27 and 188 transcripts for all the 7 bank voles, on average 66 transcripts per individual. However, for the primer design, we chose more stringent parameters to ensure retrieval of the high-quality, full-length transcripts of the classical MHC class I genes. In total, 112 unique MHC class I predicted sequences were retrieved from 5 (out of 7) individual transcriptomes. Most of the predicted sequences either possessed gaps or differed by single substitutions from a small set of unique long sequences. For simplicity, the 112 predicted sequences were grouped in 7 clusters of sequences whose members shared >90% similarity, and the alignment of representative sequences from each cluster to the mouse and rat (Rattus norvegicus) references is shown in Supplementary Figure S2. We added rat RT1-A1 sequence (1566 bp, NM_001008827.1) in the alignment to highlight the conserved regions in the transcripts, as well as to illustrate the difficulties in the alignment of the final exons.
The 112 recovered transcripts covered >90% of reference length in five sequences obtained from ‘set A’ clones (Supplementary Table S2). The ‘set A’ contained three (out of seven) samples that were used for transcriptome assembly. Two cloned sequences (5 and 6) did not have any transcript prediction that covered >80% of the reference, highlighting the low efficiency of de novo assembly in retrieving full unique sequences of polymorphic MHC genes.
Cloned transcripts of MHC class I bank vole genes and phylogenetic analysis
Sanger sequencing of clones of full-length amplicons that were obtained with the MyglMHCI_F and MyglMHCI_R primers resulted in 133 (68 from ‘set A’ and 45 from ‘set B’) sequences that showed significant similarity to predicted bank vole transcripts and rodent MHC class I sequences from GenBank. Seven unique sequences were confirmed in set A, and five in set B. Altogether, 12 sequences of the bank vole MHC class I messenger transcript were obtained (see Supplementary File S1). The seven sequences confirmed for the set A were also used to retrospectively evaluated the bank vole transcriptomic MHC class I predictions (Supplementary Figure S3). For phylogenetic analysis, 11 sequences that spanned the complete 2–4 exons (819 bp) of MHC class I were used (out of 12, as Mygl-02 and Mygl-12 were identical in this region). These sequences were analysed together with those from three other Cricetidae rodents (Chinese hamster, North American deer mouse, hispid cotton rat) and one member of the Muridae family, the house mouse. Human HLA sequences were used as an outgroup. The resulting phylogeny (Figure 2) reveals the presence of monophyletic groups at the species level, with the exception of two sequences (Pema13 and Hm1-C3) that most likely represented nonclassical (MHC Ib) genes (Crew et al., 1996).
A phylogenetic tree constructed using the maximum likelihood method (model: GTR+G+I) for exons 2–4 of MHC class I from four Cricetidae species—Myodes glareolus (Mygl-1-11, this study), Cricetulus griseus (Hm1-C2 (gi:345842483), Hm1-C4 (gi:345842478), Hm1-C5 (gi:345842476), Hm1-C3 (gi:345842485)), Peromyscus maniculatus (Pema-62 (gi:531453), Pema-41 (gi:531449), Pema-11 (gi:576635), Pema-52 (gi:531451), Pema-53 (gi:576637), Pema-13 (gi:52078311)), Sigmodon hispidus (Sihi-H2Dr, gi:19387297)—as well as Mus musculus (Muridae, H2-K1 (gi:133922587), H2-L (gi:392357526)), with Homo sapiens (HLA-A, gi: 337752171; HLA-B, gi: 170650640; HLA-C, gi: 339882742) as an outgroup. Bootstrap values based on 5000 replicates are shown. Black circles mark sequences found in ‘set A’, white circles, ‘set B’ and the grey circle represents a sequence found in both sets.
Bank vole MHC class I exon 3 genotyping and polymorphism
Semiconductor sequencing with Personal Genome Machine (PGM, Ion Torrent, Thermo Fisher Scientific, Waltham, MA, USA) of replicate spleen fragments from 29 individuals resulted in 130 563 reads (mean per individual: 4502, range: 1843–8611, s.d. 1874) for the first replicate set (I1), and 609 994 reads (mean per individual: 21 034, range: 10 753–34 161, s.d. 7176) for the second replicate set (I2). Altogether, between 54 and 95% of the demultiplexed reads were included in 31 to 60 clusters formed into individual amplicons. The remaining low-frequency, unclustered sequences and singletons were discarded. Chimeras (as detected by AmpliSAS) were present in all amplicons, with a maximum PAF of 2.22% for I1 and 2.18% for I2, and they represented 12.7% of the clustered reads.
After clustering, filtering of chimeras and manual correction of four clustering artefacts, 520 and 436 variants were assigned to I1 and I2, respectively; of these, 325 were identical in both replicates. Even though uniquely assigned variants (sequences found in only one of the two replicates) represented 48% of all assignments, they constituted only 2% of all reads, and the majority of them (85%) had a low PAF, <0.45%. They likely represented artefacts (generated by, for example, contamination, complex chimeras, multiple substitutions introduced by both PCR and sequencing and so on). In contrast, the majority (89%) of shared assignments had a frequency of >0.45% in at least one replicate. Following this observation we set an additional criterion for a variant to be accepted as a true allele: apart from being present in both replicates, it had to be present in at least one of them at a PAF of at least 0.45%. This additional threshold may have resulted in excluding alleles with low expression, but allowed us to conservatively avoid replicable artefacts. A comparison of PAFs of uniquely assigned variants and those present in both replicates is shown in Figure 3.
Our genotyping method called between 5 and 14 expressed alleles per individual (mean: 9.6, median: 10). Overall, in 29 samples, 73 different alleles were identified that translated into 69 amino acid sequences. Of these alleles, 40% were present only in one individual. The phylogenetic analysis (Supplementary Figure S4) revealed that one sequence was more divergent than the others and did not BLAST with classical MHC class I sequences (its best score was with the automatically predicted Cricetulus griseus H-2 Q10 molecule, XM_007630959.1); it therefore may represent the nonspecific amplification of a nonclassical MHC class I molecule (Ib), and was excluded from further analysis. The 72 remaining sequences were deposited in GenBank with accession numbers KU513877–KU513948, and designated Mygl-IA*01-*72. One of the deposited sequences had a 3 bp deletion in a position where the majority of the alleles possessed glycine. Final genotyping results and initial input files (after demultiplexing) are available in the Supplementary File S3.
Signatures of selection in bank vole MHC class I exon 3 sequences
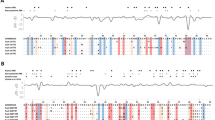
Maximum likelihood-based random-sites models from CODEML in PAML4 (Yang, 2007) were applied to 273 bp of the third exon of the MHC class I molecule that encodes amino acids 91–181 of the α2 domain (Reche and Reinherz, 2003). The results are summarised in Tables 2 and 3. The models that allowed for positive selection (M2a, M3 and M8) fit the data significantly better (P<0.001) than those that did not allow for ω>1 (M0, M1a, M7; Table 3), thus demonstrating variable selective pressure among codon positions. Although most codon positions (>90%, Table 2) were under purifying (<<1) or no apparent selection pressure (ω≈1), 8 sites (out of 91) were inferred by Bayes empirical Bayes to be under positive selection (ω>1), with posterior probability >0.95 or >0.99. Out of seven polymorphic antigen-binding sites (as defined by Reche and Reinherz, 2003 in human HLA class I), six were perfectly matched by the positively selected sites in bank vole (Figure 4).
Posterior means of ω, calculated as the average of ω over the 11 site classes, weighted by the posterior probabilities under the M8 model. The posterior probabilities were computed by the Bayes empirical Bayes procedure implemented in CODEML from the PAML4 package (Yang, 2007). Amino acid numbering is according to Reche and Reinherz (2003). Arrows indicate the polymorphic sites involved in antigen binding (Reche and Reinherz, 2003). Sites under positive selection: *P>95% and **P>99%.
Discussion
In the present study we demonstrate the usefulness of RNA-Seq de novo transcriptome assemblies in the characterisation of fast-evolving and highly polymorphic gene families. Lacking any prior knowledge about MHC class I polymorphism or copy number variation of the species of interest, we were able to use an RNA-Seq experiment to design specific primers to amplify and characterise nearly full-length genes. We then used another set of primers to investigate polymorphism and selection acting on a functionally important and highly polymorphic gene fragment (third exon) of MHC class I. The utility and accuracy of this approach can be demonstrated first by the benchmarking of the de novo assembly of the mice data set against a well-established reference and, second, by our success in the characterisation of bank vole MHC class I genes, including the obtained evidence of expected patterns of molecular evolution and fast gene turnover (as suggested by a lack of orthology to other rodent sequences).
The main conclusion that can be drawn from the benchmarking of de novo transcriptome assemblies of two mouse RNA-Seq experiments is that transcript predictions are accurate for conserved genes (like the control B2M or GADPH), but not for polymorphic and duplicated genes, such as MHC. In the case of classical MHC class I, transcript assemblies are chimeras that include partial sequences from the two loci (H2-K1 and H2-D1) of the studied mouse strain (Supplementary Figure S2). Similarly, the de novo assembled bank vole contigs never perfectly matched the transcript sequences that were obtained from the same individuals by cloning and Sanger sequencing (Supplementary Table S2). Although the majority of aligned nucleotides matched between a clone and its most similar contig, there were stretches of sequence that were likely partial chimeras of alternatively assembled contigs or paralogous transcripts, and for few nucleotide positions none of the retrieved transcripts had a matching nucleotide. It is noteworthy that in some cases the assembler failed to incorporate the correct base into the contig, even though some reads did contain the correct base, as was demonstrated by mapping the RNA-Seq reads to the sequences of cloned transcripts (Supplementary Figure S5). Thus, at present, de novo transcriptome assembly cannot be recommended as an approach for the recovery of accurate allelic sequences at a highly polymorphic and duplicated locus. Nevertheless, this finding should not discourage the use of the predicted sequences for primer design. This approach ensures specificity that is often unattainable when primer design is based on sequences from even closely related species, and thus should be a method of choice even when such resources are available. As evidenced by the benchmarking of the mouse data, conserved regions are assembled with great accuracy even in chimeric transcripts. Designing primers in the conserved regions that were identified in the bank vole transcripts allowed us to characterise the expressed MHC class I genes for the first time in this species. To the best of our knowledge, MHC class I has never been described in such detail in any Arvicolinae rodent.
Phylogenetic analysis of the exons that encoded the extracellular portion of the MHC class I molecule (domains α1, α2 and α3) that were obtained in this project and of available sequences from M. musculus and three Cricetidae species (C. griseus, P. maniculatus and S. hispidus) revealed that the MHC class I sequences found in the bank vole are more closely related to each other than to any sequence from the other species. Therefore, unless the relationships have been obscured by gene conversion (see, for example, Marková et al., 2014; see below), our findings demonstrate a lack of orthology, as found on earlier studies for MHC class I in humans and rodents (Hughes and Nei, 1989a; Crew et al., 1996). The independent origin of classical MHC class I genes in different species (that is, following the divergence of the species) was previously shown for sister subfamilies of rodents, for example, in mouse and rat (Hurt et al., 2004). These results are consistent with the faster gene turnover in MHC class I compared with MHC class II, where alleles are often shared between species and allelic lineages may be retained long after speciation. In the case of bank vole MHC class II, a trans-species polymorphism shared with a relatively closely related species from Arvicolinae was demonstrated by Bryja et al. (2006) for complete exon 2 of DQA. Evidence of sharing allelic lineages between sister subfamilies of Cricetidae was found for DQB, where one sequence of bank vole DQB clusters more closely with Peromyscus spp. than with other M. glareolus sequences (bootstrap support 60, Scherman et al., 2014). For DRB there is mixed evidence of trans-species polymorphism between subfamilies shown for exon 2 alone (Babik and Radwan, 2007), but not for longer sequences spanning exons 1–6 (Scherman et al., 2014).
In addition to retrieving nearly full-length coding sequence with traditional cloning and Sanger sequencing, we took advantage of massively parallel sequencing to comprehensively examine the polymorphism of the MHC class I third exon. We found a total of 72 alleles in 29 individuals, and this is much more than has been reported for MHC class II in the bank vole (for example, 39 alleles in 43 individuals for DQB, Scherman et al., 2014).
The frequent duplication of MHC genes is considered to be one of the driving forces in the evolution of this gene family (Yeager and Hughes, 1999). Duplication of MHC class Ia loci has been found previously in multiple rodents, for example, in Muridae (Rubocki et al., 1990; Hurt et al., 2004), Sciuridae (Kuduk et al., 2012b) and Cricetidae (Crew et al., 1990, 1996). In the mouse, the number of expressed classical genes is limited to three (but may be variable between haplotypes): H2-K1, H2-D and H2-L. The deer mouse (P. maniculatus), a member of the Cricetidae family, possesses five Pema-A loci that were identified as classical (H2-K/D/L/Q-like, Crew et al., 1996). The bank voles sampled here possessed between 5 and 14 alleles of MHC class Ia per individual, suggesting a variable number of expressed loci that range from 3 to at least 7. Allele sharing between loci, due to recent duplication and/or gene conversion (Reusch et al., 2004), could also explain the observed between-individual differences in the number of alleles. A phylogenetic tree (Supplementary Figure S4) showed no distinct clustering, preventing us from provisional assignment of the alleles to specific loci. This again suggests recent duplication, as well as possibly gene conversion and interlocus recombination, that blurs sequence differences between loci. Our finding is in line with a remark of Kulski et al. (2002) that rodents (exemplified by the mouse and the rat) exhibit a high number of duplicated MHC class I genes, greater than other studied orders of mammals (for example, primates, artiodactyls). With regards to MHC class II genes, the bank vole has also been reported as one of the species with extensive duplication, with up to eight alleles of DRB and DQB found per individual (see, for example, Axtner and Sommer 2007; Scherman et al., 2014).
Overcoming the difficulties associated with the design of reliable primers and the genotyping of complex, co-amplifying loci allowed us to examine the signatures of selection in MHC class I. Unlike for MHC class II, this has rarely been done in rodents (but see, for example, Kuduk et al., 2012b), and never in a system as complex as that found in the bank vole. We found a strong signal of positive selection in eight sites, with ω values of 3.71 (for the M8 model). Six (97, 114, 116, 152, 156 and 163) out of the eight sites that were inferred to be under positive selection perfectly match polymorphic antigen-binding sites in human HLA-A,B,C (as reported by Reche and Reinherz, 2003). Polymorphisms at antigen-binding sites affect the specificity and binding proprieties of the molecule (Reche and Reinherz, 2003), and an elevated dN/dS ratio in the α1α2 domains of MHC class I (and α1β1 in MHC class II) is interpreted to be the result of balancing selection resulting from host–-parasite interactions (Hughes and Nei, 1989a; Bernatchez and Landry, 2003). The two remaining positively selected bank vole sites (150 and 155) are located in the proximity of the antigen-binding residues, and are potentially involved in the interaction with T-cell receptor Vα and Vβ chains (Reche and Reinherz, 2003).
In conclusion, this study demonstrates the suitability of de novo transcriptome assemblies for the study of highly polymorphic and fast-evolving gene families, in particular for PCR primer development. Our study adds new data to the so-far limited collection of MHC class I sequences from nonmodel species that broadens the scope for comparative studies of rodent and mammalian immune systems. Our analyses support the fast rate of evolution of MHC class I in the bank vole. Interestingly, although no apparent orthology to sequences from other subfamilies of Muridae rodents was shown, the amino acid sites under positive selection are nonetheless shared among mammals. Resources from our study will be highly relevant to surveys that use the bank vole as a model species in ecology and evolution. In particular, they will facilitate studies of the association of MHC class I variation with pathogen infection, for example, susceptibility to the Puumala virus that infects bank voles in many regions of Europe and constitutes a health threat to humans.
Data archiving
Sequences retrieved with cloning and Sanger sequencing of full MHC I transcripts are available in Supplementary File S1. Demultiplexed reads from HTS experiment and final genotyping results are in the Supplementary File S3. Allele sequences after genotyping of MHC I exon 3 have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank/): accession numbers KU513877–KU513948. Tools used in the genotyping of MHC class I sequences are accessible at: http://evobiolab.biol.amu.edu.pl/amplisat/.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Apanius V, Penn D, Slev PR, Ruff LR, Potts WK . (1997). The nature of selection on the major histocompatibility complex. Crit Rev Immunol 17: 179–224.
Axtner J, Sommer S . (2007). Gene duplication, allelic diversity, selection processes and adaptive value of MHC class II DRB genes of the bank vole, Clethrionomys glareolus. Immunogenetics 59: 417–426.
Babik W . (2010). Methods for MHC genotyping in non-model vertebrates. Mol Ecol Resour 10: 237–251.
Babik W, Radwan J . (2007). Sequence diversity of MHC class II DRB genes in the bank vole Myodes glareolus. Acta Theriol 52: 227–235.
Babik W, Taberlet P, Ejsmond MJ, Radwan J . (2009). New generation sequencers as a tool for genotyping of highly polymorphic multilocus MHC system. Mol Ecol Resour 9: 713–719.
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J et al. (2013). GenBank. Nucleic Acids Res 41: D36–D42.
Bernatchez L, Landry C . (2003). MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol 16: 363–377.
Bragg LM, Stone G, Butler MK, Hugenholtz P, Tyson GW . (2013). Shining a light on dark sequencing: characterising errors in Ion Torrent PGM data. PLoS Comput Biol 9: e1003031.
Bryja J, Galan M, Charbonnel N, Cosson JF . (2006). Duplication, balancing selection and trans-species evolution explain the high levels of polymorphism of the DQA MHC class II gene in voles (Arvicolinae). Immunogenetics 58: 191–202.
Crew MD, Bates LM, Douglass CA, York JL . (1996). Expressed Peromyscus maniculatus (Pema) MHC class I genes: evolutionary implications and the identification of a gene encoding a Qa1-like antigen. Immunogenetics 44: 177–185.
Crew MD, Filipowsky ME, Neshat MS, Smith GS, Walford RL . (1991). Transmembrane domain length variation in the evolution of major histocompatibility complex class I genes. Proc Natl Acad Sci USA 88: 4666–4670.
Crew MD, Filipowsky ME, Zeiler EC, Smith GS, Walford RL . (1990). Major histocompatibility complex class I genes of Peromyscus leucopus. Immunogenetics 1: 371–379.
Darriba D, Taboada GL, Doallo R, Posada D . (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772.
Deter J, Bryja J, Chaval Y, Galan M, Henttonen H, Laakkonen J et al. (2008). Association between the DQA MHC class II gene and Puumala virus infection in Myodes glareolus, the bank vole. Infect Genet Evol 8: 450–458.
Ejsmond MJ, Radwan J . (2015). Red Queen processes drive positive selection on major histocompatibility complex (MHC) genes. PLoS Comput Biol 11: e1004627.
Filipi K, Marková S, Searle JB, Kotlík P . (2015). Mitogenomic phylogenetics of the bank vole Clethrionomys glareolus, a model system for studying end-glacial colonization of Europe. Mol Phylogenet Evol 82 (Pt A): 245–257.
Gourraud P-A, Khankhanian P, Cereb N, Yang SY, Feolo M, Maiers M et al. (2014). HLA diversity in the 1000 Genomes dataset. PLoS One 9: e97282.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson D a, Amit I et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652.
Guindon S, Gascuel O . (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704.
Guivier E, Galan M, Salvador AR, Xuéreb A, Chaval Y, Olsson GE et al. (2010). Tnf-α expression and promoter sequences reflect the balance of tolerance/resistance to Puumala hantavirus infection in European bank vole populations. Infect Genet Evol 10: 1208–1217.
Hughes AL, Nei M . (1989a). Evolution of the major histocompatibility complex: independent origin of nonclassical class I genes in different groups of mammals. Mol Biol Evol 6: 559–579.
Hughes AL, Nei M . (1989b). Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA 86: 958–962.
Hurt P, Walter L, Sudbrak R, Klages S, Müller I, Shiina T et al. (2004). The genomic sequence and comparative analysis of the rat major histocompatibility complex. Genome Res 14: 631–639.
Hutchins AP, Poulain S, Fujii H, Miranda-Saavedra D . (2012). Discovery and characterization of new transcripts from RNA-seq data in mouse CD4(+) T cells. Genomics 100: 303–313.
Jeffery KJM, Bangham CRM . (2000). Do infectious diseases drive MHC diversity? Microbes Infect 2: 1335–1341.
Kalendar R, Lee D, Schulman AH . (2014). FastPCR software for PCR, in silico PCR, and oligonucleotide assembly and analysis. Methods Mol Biol 1116: 271–302.
Katoh K . (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066.
Katoh K, Standley DM . (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780.
Kelley J, Walter L, Trowsdale J . (2005). Comparative genomics of major histocompatibility complexes. Immunogenetics 56: 683–695.
Klein J . (1986) Natural History of the Major Histocompatibility Complex. Wiley: New York.
Kloch A, Babik W, Bajer A, Siński E, Radwan J . (2010). Effects of an MHC-DRB genotype and allele number on the load of gut parasites in the bank vole Myodes glareolus. Mol Ecol 19 (Suppl 1): 255–265.
Konczal M, Koteja P, Orlowska-Feuer P, Radwan J, Sadowska ET, Babik W . (2016). Genomic response to selection for predatory behavior in a mammalian model of adaptive radiation. Mol Biol Evol 33: 2429–2440.
Kotlík P, Marková S, Vojtek L, Stratil A, Slechta V, Hyršl P et al. (2014). Adaptive phylogeography: functional divergence between haemoglobins derived from different glacial refugia in the bank vole. Proc Biol Sci 281: 20140021.
Kuduk K, Babik W, Bojarska K, Sliwińska EB, Kindberg J, Taberlet P et al. (2012a). Evolution of major histocompatibility complex class I and class II genes in the brown bear. BMC Evol Biol 12: 197.
Kuduk K, Johanet A, Allainé D, Cohas A, Radwan J . (2012b). Contrasting patterns of selection acting on MHC class I and class II DRB genes in the Alpine marmot (Marmota marmota). J Evol Biol 25: 1686–1693.
Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H . (2002). Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev 190: 95–122.
Langmead B, Salzberg SL . (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359.
Li B, Fillmore N, Bai Y, Collins M, Thomson JA, Stewart R et al. (2014). Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol 15: 553.
Lighten J, van Oosterhout C, Bentzen P . (2014). Critical review of NGS analyses for de novo genotyping multigene families. Mol Ecol 23: 3957–3972.
Miller M, Kaukinen KH, Angel K, Kaukinen KH, Schulze AD . (2002). Expansion and contraction of major histocompatibility complex genes: a teleostean example. Immunogenetics 53: 941–963.
Marková S, Filipi K, Searle JB, Kotlík P . (2015). Mapping 3’ transcript ends in the bank vole (Clethrionomys glareolus) mitochondrial genome with RNA-Seq. BMC Genomics 16: 870.
Marková S, Searle JB, Kotlík P . (2014). Relaxed functional constraints on triplicate α-globin gene in the bank vole suggest a different evolutionary history from other rodents. Heredity (Edinb) 113: 64–73.
McElroy KE, Luciani F, Thomas T . (2012). GemSIM: general, error-model based simulator of next-generation sequencing data. BMC Genomics 13: 74.
Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L et al. (2013). Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14: 193–202.
Mokkonen M, Kokko H, Koskela E, Lehtonen J, Mappes T, Martiskainen H et al. (2011). Negative frequency-dependent selection of sexually antagonistic alleles in Myodes glareolus. Science 334: 972–974.
Rada C, Lorenzi R, Powis SJ, van den Bogaerde J, Parham P, Howard JC . (1990). Concerted evolution of class I genes in the major histocompatibility complex of murine rodents. Proc Natl Acad Sci USA 87: 2167–2171.
Radwan J, Zagalska-Neubauer M, Cichoń M, Sendecka J, Kulma K, Gustafsson L et al. (2012). MHC diversity, malaria and lifetime reproductive success in collared flycatchers. Mol Ecol 21: 2469–2479.
Reche PA., Reinherz EL . (2003). Sequence variability analysis of human class I and class II MHC molecules: Functional and structural correlates of amino acid polymorphisms. J Mol Biol 331: 623–641.
Reusch TBH, Schaschl H, Wegner KM . (2004). Recent duplication and inter-locus gene conversion in major histocompatibility class II genes in a teleost, the three-spined stickleback. Immunogenetics 56: 427–437.
Rice P, Longden I, Bleasby A . (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277.
Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SGE . (2013). The IMGT/HLA database. Nucleic Acids Res 41: D1222–D1227.
Rubocki RJ, Lee DR, Lie WR, Myers NB, Hansen TH . (1990). Molecular evidence that the H-2D and H-2L genes arose by duplication. Differences between the evolution of the class I genes in mice and humans. J Exp Med 171: 2043–2061.
Sadowska ET, Baliga-Klimczyk K, Chrzaścik KM, Koteja P . (2008). Laboratory model of adaptive radiation: a selection experiment in the bank vole. Physiol Biochem Zool 81: 627–640.
Scherman K, Råberg L, Westerdahl H . (2014). Positive selection on MHC class II DRB and dqb genes in the bank vole (Myodes glareolus). J Mol Evol 78: 293–305.
Sebastian A, Herdegen M, Migalska M, Radwan J . (2016). amplisas: a web server for multilocus genotyping using next-generation amplicon sequencing data. Mol Ecol Resour 16: 498–510.
Sommer S, Courtiol A, Mazzoni CJ . (2013). MHC genotyping of non-model organisms using next-generation sequencing: a new methodology to deal with artefacts and allelic dropout. BMC Genomics 14: 542.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S . (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729.
Vaheri A, Henttonen H, Voutilainen L . (2013). Hantavirus infections in Europe and their impact on public health. Rev Med Virol 23: 35–49.
Westerdahl H, Waldenström J, Hansson B, Hasselquist D, von Schantz T, Bensch S . (2005). Associations between malaria and MHC genes in a migratory songbird. Proc Biol Sci 272: 1511–1518.
Yang Z . (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591.
Yang Z, Wong WSW, Nielsen R . (2005). Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22: 1107–1118.
Yeager M, Hughes A . (1999). Evolution of the mammalian MHC: natural selection, recombination, and convergent evolution. Immunol Rev 167: 45–58.
Zagalska-Neubauer M, Babik W, Stuglik M, Gustafsson L, Cichoń M, Radwan J . (2010). 454 sequencing reveals extreme complexity of the class II major histocompatibility complex in the collared flycatcher. BMC Evol Biol 10: 395.
Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P . (2011). Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics 12 (Suppl 1): S2.
Zieliński P, Stuglik MT, Dudek K, Konczal M, Babik W . (2014). Development, validation and high-throughput analysis of sequence markers in nonmodel species. Mol Ecol Resour 14: 352–360.
Acknowledgements
We sincerely thank Paweł Koteja for the donation of bank vole spleens and Michał Kolasa for the collection of vole tissues during necropsy. We thank Sylwia Jedut, Julia Olechnowicz and Silvia Marková for assistance in the laboratory. This work was supported by Grant UMO-2013/08/A/NZ8/00153 from the National Science Centre (NCN) to J Radwan and by Grants from the Czech Science Foundation (P506-11-1872 and 16-03248S) to P Kotlík.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Rights and permissions
About this article
Cite this article
Migalska, M., Sebastian, A., Konczal, M. et al. De novo transcriptome assembly facilitates characterisation of fast-evolving gene families, MHC class I in the bank vole (Myodes glareolus). Heredity 118, 348–357 (2017). https://doi.org/10.1038/hdy.2016.105
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2016.105
This article is cited by
-
Characterization of two MHC II genes (DOB, DRB) in white-tailed deer (Odocoileus virginianus)
BMC Genetics (2020)
-
Profiling of the TCRβ repertoire in non-model species using high-throughput sequencing
Scientific Reports (2018)