Abstract
Furunculosis (Aeromonoas salmonicida) is an important disease in Atlantic salmon (Salmo salar) farming. Vaccination and selective breeding for increased resistance to the disease on the basis of challenge tests of unvaccinated fish are used as complementary prophylactic methods. An important issue is whether genetic predisposition to infection is consistent across vaccinated and unvaccinated fish. Hence, the main objective of this study was to determine the magnitude of the genetic associations (correlations) between resistance to furunculosis in vaccinated and unvaccinated fish, and to estimate the magnitude of the correlation of resistance to furunculosis with resistance to the viral diseases infectious pancreatic necrosis (IPN) and infectious salmon anaemia (ISA). Sub-samples of unvaccinated and vaccinated salmon from 150 full-sib families were subjected to separate cohabitation challenge tests. Substantial genetic variation was found in resistance to furunculosis in both the unvaccinated (heritabilities of 0.51±0.05) and vaccinated (0.39±0.06) fish. However, the genetic correlation between resistance to furunculosis in the two groups was low (0.32±0.13), indicating a weak genetic association between resistance in the two groups. Hence, the current selection strategy on the basis of challenge tests of unvaccinated fish is likely to produce low genetic improvement in resistance to furunculosis under field conditions, where fish are vaccinated with an effective vaccine. Evidence was found of significantly favourable genetic associations of resistance to furunculosis in unvaccinated (but less so for vaccinated) fish with resistance to both IPN and ISA (unvaccinated fish), indicating that vaccination ‘mask’ genetic associations between resistance to different diseases.
Similar content being viewed by others
Introduction
Vaccination is important in preventing outbreaks of infectious diseases (NOHA, 2010), both for humans and livestock. In farmed fish, especially salmonids, vaccination has been the single most efficient prophylactic measure and has contributed to a significant reduction in the use of antibiotics in the Norwegian salmon industry in the 1990's (Ellis, 1997; Gudding et al., 1999; Håstein et al., 2005) Currently, farmed Atlantic salmon are routinely vaccinated as pre-smolts against a number of diseases, both bacterial (furunculosis, vibriosis, cold water vibriosis, winter ulcer) and viral (infectious pancreatic necrosis (IPN), pancreas disease, infectious salmon anaemia (ISA)).
At the same time, several studies have provided ample evidence that there is genetic variation in resistance against bacterial and viral diseases in Atlantic salmon. For example, heritability estimates for survival in challenge tests range from 0.43 to 0.62 for Aeromonas salmonicida (subsp. salmonicida), the causative agent of furunculosis (Gjedrem et al., 1991; Kjøglum et al., 2008; Ødegård et al., 2007b). These findings have motivated breeding companies to include disease resistance in their breeding goals (Kjøglum et al., 2008). However, genetic variation in disease resistance has been based on unvaccinated fish and we were thus interested in examining to what extent the same genetic traits are involved in resistance to challenge in vaccinated and unvaccinated fish. We hypothesized that different parts of the immune system have a role in protection against disease in vaccinated and unvaccinated fish. Through vaccination, the fish have mounted an immune response before infection, which may reduce the relative importance of the innate defence mechanisms, although anatomical barriers and innate immune responses are likely to have a role also in the vaccinated fish. The details of immune responses to vaccination have not been studied in fish, nor are they well understood in higher vertebrates (Nakayama et al., 2010). Hence, the genetics underlying disease resistance may be quite different in the two situations, and to date, there are no data available to determine if genetic improvement of disease resistance on the basis of unvaccinated fish would also lead to enhanced disease resistance in vaccinated fish (beyond the effect of vaccination).
In terrestrial farmed species, estimates of genetic variation in the response to an infectious disease are generally based on individual field records, where the exposure to the pathogen can be incomplete and often result in low frequency of the disease (Bishop and Woolliams, 2010). In aquaculture species, however, challenge tests exposing all individuals to the infection can be performed by either intraperitoneal injection (i.p.), bath or cohabitation challenge tests. The latter method infect the test fish through the natural route of infection (Nordmo, 1997), and the genetic correlation for survival tested by experimental challenge and in the field was found to be close to unity (0.95) for furunculosis (Gjøen et al., 1997) and also high (0.78–0.83) for IPN (a double-stranded RNA virus) (Wetten et al., 2007).
Furunculosis was the first disease to be included in the breeding goal for Atlantic salmon. Currently, an effective vaccine is available for this disease, thus representing a valuable model to study disease resistance in vaccinated and unvaccinated animals. The main objective of this study was to estimate the genetic association of protection against a bacterial disease (furunculosis) in unvaccinated and vaccinated Atlantic salmon (including two different vaccines). The secondary aim was to estimate the genetic associations between resistance against furunculosis and two viral diseases (ISA and IPN) currently included in breeding programs for Atlantic salmon.
Materials and methods
Family material
The fish in this study were all from the breeding nucleus population of SalmoBreed AS. In total, 279 full-sib families (offspring of 140 sires and 279 dams) were included in the analysis. The families were produced by Bolaks, Eikelandsosen, Norway in November 2006. In January 2007, they were transported as eyed eggs to Nofima Marin, Sunndalsøra, Norway, where they were reared in separate trays. From first feeding (5 February to 17 April 2007), they were kept in separate 0.75 m3 tanks, until they were tagged with PIT (Passive Integrated Transponder) tags in July and September 2007 (Table 1). The environmental conditions were standardized during the hatchery and rearing period until tagging to minimize environmental differences between families. Additive genetic relationships were available between the individuals, as well as among their parents.
In the present population, selection for increased resistance against furunculosis on the basis of challenge test survival data (with unvaccinated fish) had been performed for one generation. The breeding goal also included resistance against ISA, maximum growth, minimum fillet fat and improved fillet colour.
Five fish groups were included in this study; two groups from a random sample of 150 of the 279 families (groups 1 and 2; the offspring of 87 sires and 150 dams; as considered adequate for estimation of genetic parameters) and three groups from all the 279 families (groups 3, 4 and 5; as they were part of the standard genetic evaluation programme of SalmoBreed AS; Bolaks). The different groups of fish all consisted of a random sample of 15 fish per family and were tested as follows:
Group 1 (Fur-SV): Fish vaccinated with a commercial multi-component vaccine and challenged with A. salmonicida.
Group 2 (Fur-RD): Fish vaccinated with an experimental multi-component vaccine with a reduced antigen dose and challenged with A. salmonicida.
Group 3 (Fur): Unvaccinated fish challenged with A. salmonicida.
Group 4 (ISA): Unvaccinated fish challenged with ISA virus.
Group 5 (IPN): Unvaccinated fish challenged with IPN virus.
The Fur-SV and Fur-RD groups were tagged in July 2007 and the Fur, ISA and IPN groups were tagged in September 2007.
Vaccines and vaccination
The Fur-SV and Fur-RD fish were vaccinated at the pre-smolt stage from 2 October to 4 October 2007. The Fur-SV fish were i.p. injected with 0.1 ml of the commercial vaccine ALPHA JECT 6–2 (AJ6–2) (PHARMAQ, Oslo, Norway). AJ6–2 includes antigens of A. salmonicida, Vibrio anguillarum serotype O1 and O2, Vibrio salmonicida, M. viscosa and IPN virus, and protects against furunculosis, classical vibriosis, cold-water vibriosis, winter ulcer and IPN, respectively. The Fur-RD fish were vaccinated with an experimental AJ6–2 of 0.05 ml per fish, containing 60% less A. salmonicida antigen than the commercial AJ6–2 used in the Fur-SV group.
The vaccines were prepared by PHARMAQ AS and are water-in-oil emulsions, where the dispersal water phase contains the inactivated viral and bacterial antigens. A reduced antigen dose of A. salmonicida in the vaccine given to the Fur-RD fish was used for two reasons: first, to see if fish given a reduced antigen dose showed different genetic variation in survival compared with fish given the commercial AJ6–2, and second, to ensure mortality in at least some vaccinated fish post-challenge. Individual body weights of the vaccinated fish were recorded at the time of vaccination.
Challenge tests
Fur group, ISA group and IPN group
Pre-smolts of the Fur-SV and Fur-RD groups were transported from Sunndalsøra to VESO Vikan (Namsos, Norway) on 15 November 2007, and the challenge test started on 22 November 2007. The Fur-SV and Fur-RD groups were challenged in the same tank, containing 3 m3 of 12 °C freshwater at an exchange rate of approximately 0.8 l kg−1 fish. The challenge was carried out by cohabitation, where naive Atlantic salmon were injected i.p., with the respective pathogens to act as shedders. The number of shedders was 20% of the total number of fish in the tank and they were added at three different time points to prevent the mortality from plateauing, before 50% control mortality was reached; 29% of the total number of shedders were added at day one of test (i.p. injected with 5.0 × 103 cfu of A. salmonicida), 29% at day five (i.p. injected with 3.0 × 101 cfu of A. salmonicida) and the remaining (42%) at day 32 (i.p. injected with 3.0 × 101 cfu of A. salmonicida). A total of 100 unvaccinated fish were randomly sampled from the 150 Fur-SV/Fur-RD families, ink tagged and used as an unvaccinated control to monitor the challenge pressure.
Fur group
Pre-smolts were transported from Sunndalsøra to VESO Vikan on 29 September 2007, and the challenge test started at 2 October 2007. All fish were tested in a single test tank, containing 3 m3 of 12 °C freshwater at an exchange rate of about 0.8 l kg−1 fish. In this test, shedders (i.p. injected with 5.0 × 103 cfu of A. salmonicida) comprising 7% of the total number of fish were used, all added on the first day of the test.
ISA group
Pre-smolts were transported from Sunndalsøra to VESO Vikan on 29 September 2007, and the challenge test started on 5 October 2007. Fish were tested in a single test tank, containing 3 m3 of 12 °C freshwater at an exchange rate of about 0.8 l kg−1 fish. In this test, shedders (i.p. injected with 6.4 × 103 TCID50 of ISA virus comprising 24% of the total number of fish were all added on the first day of the test.
IPN group
Post-smolts were transported from Sunndalsøra to VESO Vikan on 21 January 2008. The fish were transferred to seawater the day after arrival, and the challenge test started on 25 January 2008. Because of a larger biomass, fish were randomly allocated to two different tanks, each of 5 m3 of 12 °C seawater, with an exchange rate set to give a minimum oxygen level of 5–6 mg O2/l in the outlet water. In this test, shedders (i.p. injected with 0.4 ml solution with approximately 1 × 107 TCID50/ml of IPN virus) comprising 28% of the total number of fish were used. The shedders were added at two time points: 50% on day one and 50% on day five.
Dead fish were recorded daily in all tests. Mortality diagnostics were performed in order to verify the causative agent of mortality. Bacteriological tests were used to detect A. salmonicida, clinical signs were used to diagnose ISA and an enzyme linked immunosorbent assay was used to detect IPN virus.
Data recording
Survival in challenge tests was defined as a binary trait, where fish that died during the test were assigned a score of zero, and fish that were still alive at the end of the test were assigned a score of one. Survival records of the different groups were treated as different traits. The challenge tests for groups Fur, ISA and IPN were terminated at intermediate mortalities (day 21, 33 and 30 of the challenge test, respectively). This has been the standard procedure for such tests as the phenotypic variance for the observed binary trait is at its maximum at this point. The challenge tests for the groups Fur-SV and Fur-RD were terminated when mortalities had essentially ceased (day 60).
Statistical analysis
Models using survival at end of test were chosen. More complex longitudinal survival models (using test-day survival) were tested, but did not give significantly different genetic associations between the disease traits (results not shown). Earlier studies have shown that simple cross-sectional models and more advanced longitudinal models give very similar ranking of families for this type of data (Gitterle et al., 2006; Ødegård et al., 2007a), which was consistent with our results.
To find the most appropriate model, linear single trait sire-dam models were first run for the traits Fur-SV, Fur-RD, Fur, ISA, IPN and body weight at vaccination. The full univariate model for each trait was:

where: yijkl=survival or body weight (trait i) for fish j, progeny of sire k and dam l, μi=the overall mean, xj=tank in challenge for fish j (only for IPN-group) bi=fixed regression coefficient of age, sirek=random additive genetic effect of sire k, daml=random additive genetic effect of dam l, familyl=random effect common to full sibs of dam l (only for body weight at vaccination), ej=random residual for fish j.
Likelihood ratio tests (Lynch and Walsh, 1998) were used to determine whether the family effect was significant in addition to the additive genetic sire and dam effects, that is, whether one should use the full or reduced model. Body weight at vaccination was the only trait where family effect were significant (P>0.05), and the family effect was thus only included for this trait in the further multi-trait models. For the software used, likelihood ratio tests can not be used for threshold models, but testing the effect of including a family effect on a linear, single trait model serves as a good approximation, even for binary traits.
Then a multi-trait linear model, including the body weight trait and the five binary disease traits (Fur-SV, Fur-RD, Fur, ISA, IPN), was run to obtain estimates of variance components on the observed linear scale for all traits. In addition, a similar multi-trait model, including a linear model for the body weight trait and a threshold (probit) model for each of the five binary traits, was used to obtain estimates of variance components on the underlying liability scale for the binary traits and covariance components among the traits. The model for each binary trait can be written as:

where Sijkl=survival trait i for fish j, of sire k, dam l,Φ( )=cumulative standard normal distribution, and the other parameters are as described above.

where u is a vector of sire and dam effects and e is the residual vector for all six traits, A is the additive relationship matrix among the sire and dams, I is an identity matrix of appropriate size, G is the additive genetic (co)variance matrix and R is the residual variance-covariance matrix among the traits, defined as:

As the last five traits were binary, recorded on different fish and fitted by a probit threshold model, the lower 5 × 5 corner of R was restricted to be an identity matrix. Furthermore, vaccine weights were only measured in vaccinated fish. Hence, only three parameters of the R matrix needed to be estimated: σ2weight,(residual variance for weight), σweight, Fur−SV (residual covariance between weight and Fur-SV) and σweight, Fur−RD (residual covariance between weight and Fur-RD).
The DMU software (Madsen and Just 2007) was used in all statistical analyses.
Heritability
For all traits, heritability was calculated as:

where; σμ2 is the additive genetic sire-dam variance (1/4 of the total additive genetic variance), σc2 is the common family variance (non-significant and therefore set to zero for all survival traits) and σe2 is the (underlying) residual variance (per definition set to 1.0 for all binary traits).
Results
Descriptive statistics
Table 1 contains descriptive statistics for body weight (at tagging and vaccination) and age (days from first feeding) of fish in the five groups. At the start of the challenge tests, the average weight of fish (based on two subsamples of 50 fish) was 46 g for the Fur-SV/Fur-RD group, 30 g for each of the Fur and ISA groups and 85 g for the IPN group.
Challenge tests survival curves
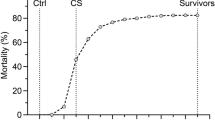
Survival curves for the two vaccinated fish groups and the unvaccinated control fish group (furunculosis) are shown in Figure 1a, and the survival curves for the three unvaccinated fish groups (furunculosis, IPN and ISA) are shown in Figure 1b. For the Fur-SV and Fur-RD groups, onset of mortality started at day 12 post challenge and remained relatively low, although higher for Fur-RD than for Fur-SV. By day 35–40, daily mortality for both these groups plateaued and the test was terminated on day 60. At the end of the test, Fur-SV showed an approximately 10% higher survival than Fur-RD (61 versus 51 %). For the unvaccinated Fur group, onset of mortality was at day eight post challenge and continued at a high rate until the test was terminated on day 21, giving a survival of 28% (Figure 1b). For the ISA group, the first mortalities were observed on day 22 post challenge and continued at a high rate until the test was terminated on day 33 at 36% survival (Figure 1b). For the IPN group, mortality started on day 12 and daily mortalities continued at an intermediate rate until the test was terminated on day 39 post challenge, resulting in 55% survival (Figure 1b).
(a) Survival curves for the two vaccinated fish groups (Fur-SV (▪) and Fur-RD (—)) and for the unvaccinated control group (Control (---)) during challenge with A. salmonicida in the same tank. (b)Survival curves for the three unvaccinated fish groups (Fur (▴), ISA (•) and IPN (x) during challenge with A. salmonicida, ISA virus and IPN virus. Challenges were carried out in three different tanks.
Heritabilities and genetic correlations
The estimated heritabilities (observed and underlying scale) are shown in Table 2. Estimated underlying heritabilities of Fur-SV and Fur-RD were high (0.39±0.06 and 0.34±0.06, respectively), but clearly lower than for the unvaccinated Fur group (0.51±0.05). Estimated underlying heritabilities for ISA and IPN (0.33±0.05 and 0.39±0.05, respectively) were similar to the heritabilities for Fur-SV and Fur-RD. As expected, the estimated heritabilities on the observed scale (obtained with a linear model) were generally lower than those on the underlying liability scale. The estimated heritability for body weight at vaccination was relatively high (h2=0.29±0.09) with a significant, albeit small, environmental effect common to full-sibs (0.09±0.09, P<0.05).
Estimated genetic correlations between the five survival traits and body weight at vaccination are shown in Table 3. All genetic correlations among survival traits (except between furunculosis in vaccinated fish and IPN) were significantly larger than zero and thus favourable. The highest genetic correlation (0.90±0.06) was between survival and furunculosis in the two vaccinated groups (Fur-SV and Fur-RD), whereas genetic correlations between survival and furunculosis in vaccinated (Fur-SV and Fur-RD) and unvaccinated (Fur) fish were medium to low (0.32–0.54), and of the same magnitude as the genetic correlations among different diseases in unvaccinated fish (Fur, ISA and IPN). The estimated genetic correlations between body weight at vaccination and the survival traits were all positive, but low and not significantly different from zero (P>0.05).
Discussion
The results in this study strongly suggest that heritability of resistance against furunculosis decreases as a result of vaccination. Hence, resistance against furunculosis that is induced by vaccination appears to ‘mask’ some of the innate genetic resistance against the disease. However, the heritability estimates for survival to furunculosis were relatively high and accurate, suggesting that there is substantial additive genetic variation in resistance against furunculosis in both unvaccinated and vaccinated fish, which can be used to improve resistance to furunculosis in Atlantic salmon through selective breeding. For the unvaccinated fish, the heritability estimates for the three studied diseases are of the same magnitude as those reported in previous studies (Ødegård et al., 2007b; Kjøglum et al., 2008; Gjerde et al., 2009; Guy et al., 2009). To our knowledge, estimates of heritability for disease resistance in vaccinated farmed fish or livestock have not been published earlier.
For the two vaccinated groups (Fur-SV and Fur-RD) the heritabilities for survival to furunculosis were very similar suggesting that the experimental vaccine, which provided lower protecting compared the commercial vaccine, was sufficient to ‘mask’ innate genetic resistance to furunculosis to the same degree as the commercial vaccine. This is also confirmed by the high genetic correlation (rg=0.90) between resistant to furunculosis in the two groups, implying that the genetic resistance is largely the same trait irrespective of the injection volume and the antigen dose of A. salmonicida in the vaccines. In contrast, the genetic correlation between survival of vaccinated and unvaccinated fish was positive, but moderate to low (rg=0.32–0.54), and suggests that resistance in vaccinated and unvaccinated fish should be regarded as partly different genetic traits. These results disagree with the results from a study of Marek's disease in chickens, where a linear relationship was observed between disease resistance in unvaccinated and vaccinated chickens from seven genetically different strains (not fullsib families as in the present study) (Gavora et al., 1990).
Our results suggest that selection for improved genetic resistance to furunculosis on the basis of testing of unvaccinated animals will also lead to some degree of improved disease resistance in vaccinated animals (the present situation in the industry). However, the moderate genetic correlation between survival in vaccinated and unvaccinated fish raises the question if the testing procedure agrees with the breeding goal for the trait resistance to furunculosis. And for such a trait the issue of short- or long-term breeding goal is of particular interest and needs to be clarified. The current challenge testing procedure using unvaccinated fish is only optimal if the long-term breeding goal is to produce a resistant or tolerant fish without vaccination. This selection strategy will also provide some increased resistance in vaccinated fish through indirect selection, but to a large extent, dependent on the economic weight given to the trait relative to other traits selected for. The most likely scenario is, however, that vaccination has to be practised by the industry in the foreseeable future (short to medium term). Therefore, selection for improved survival based on testing of vaccinated fish would be more efficient and relevant for the salmon farming industry today.
Differences in challenge methods (duration) may also have affected the genetic correlation between resistance to furunculosis in vaccinated and unvaccinated fish. The unvaccinated fish in this study were tested using a standard challenge test procedure (SalmoBreed breeding program), where the test is terminated at an intermediate and still high mortality, whereas the vaccinated fish groups were tested over a much longer time period (with repeated challenges using additional shedders) and the test terminated when mortality had essentially ceased. End-survival variation may be explained by differences in susceptibility to the disease (given that the population contains a fraction of non-susceptible fish) and variation in individual hazard rates (time until death), given that the fish are susceptible. These two traits may be governed by differing genetic factors, as has previously been estimated in challenge tests with the ectoparasite Gyrodactylus salaris in Atlantic salmon (Salte et al., 2010) and taura syndrome virus in Pacific white shrimp (Penaeus vannamei) (Ødegård et al., 2011). In this case, the relative importance of susceptibility status and hazard rate (given susceptibility), with respect to final survival, will depend on the chosen endpoint of the test. Generally, earlier termination of the test (at a still high mortality) will increase the relative importance of individual hazard rates and decrease the relative importance of susceptibility. In addition, early termination of the test can potentially lower the specificity, in the sense that some non-healthy individuals will be classified as healthy (survivors) even though they would have died given sufficient challenge time. Low specificity can often be seen in field data due to incomplete exposure to the pathogen (Bishop and Woolliams, 2010), whereas challenge tests aim to expose all individuals to the specific infection. Exposure to the pathogen is needed for an individual to express its genotype for resistance, and in the challenge test with vaccinated fish in this study, shedders were added several times to ensure this. In challenge tests with unvaccinated fish, the disease will spread faster and shedders were thus only added at the first day of challenge. In the present study, all families were kept in the same tank in each challenge test, and a genetic correlation close to unity (0.95) has been estimated between survival to furunculosis in challenge and field test with unvaccinated fish (Gjøen et al., 1997). It cannot be ruled out that differences in pathogenic pressure may have some effect on ranking of families, but performing the test in replicated tank was not possible due to financial constraint as such tests are expensive to perform.
The genetic correlation between resistance to furunculosis and ISA in unvaccinated fish was moderately positive and is concordant with results from Ødegård et al. (2007b), who reported a positive, but lower genetic correlation between the two traits. Conversely, other studies have found this genetic correlation to be not significantly different from zero (Kjøglum et al., 2008), or low and even negative (Gjøen et al., 1997). In all previous studies, ISA virus challenges were partly or exclusively performed using i.p. injections, whereas cohabitation challenges were used for A. salmonicida. In the current study, cohabitation challenge was used for both furunculosis and ISA. Cohabitation challenge, which mimics the natural route of infection, will provide a better estimate of the genetic variation underlying protective responses, whereas i.p. injections, where natural barriers are surpassed, will likely reveal a restricted profile of the underlying genetic variation. Estimates of positive genetic correlations across diseases may be due to the innate defence mechanisms and are therefore likely more precise when the pathogen exposure mimics natural infection as close as possible. In contrast, such a correlation may be less obvious using artificial pathogen exposure, for example, through the i.p. route. The challenge model is thus likely to influence the magnitude of the genetic correlations between resistance to different diseases.
The heritability estimate for IPN was intermediate (0.39±0.05) and is concordant with previous studies (Wetten et al., 2007; Kjøglum et al., 2008). A quantitative trait loci explaining a very large proportion of the genetic variation in IPN resistance has been detected in Atlantic salmon (Houston et al., 2009; Moen et al., 2009). In our study, evidence was found of significantly favourable genetic correlations of resistance to furunculosis in unvaccinated fish with resistance to both IPN and ISA (unvaccinated fish). Kjøglum et al. (2008) reported a non-significant genetic correlation between these two traits, but i.p. injections and immersions were used as challenge models and IPN resistance was tested using fry and not pre-smolt. In our study, the correlations between IPN and ISA were lower for the vaccinated fish groups, indicating that vaccination (in this case against furunculosis) ‘mask’ genetic associations between resistant to different diseases.
The present findings indicate that genetic resistance against furunculosis in vaccinated and unvaccinated fish is controlled by partially different genetic mechanisms, thus questioning selective breeding for furunculosis resistance in vaccinated fish based on testing of unvaccinated fish. Furthermore, potential compromises between vaccination programs and selective breeding programs for enhanced resistance against infectious diseases should be investigated for other diseases, where vaccines are widely used.
References
Bishop SC, Woolliams JA (2010). On the genetic interpretation of disease data. PLoS ONE 5: e8940.
Ellis AE (1997). Immunization with bacterial antigens: Furunculosis. Dev Biol Stand 90: 107–116.
Gavora JS, Spencer JL, Okada I, Grunder AA (1990). Correlation of genetic-resistance of chickens to Mareks-disease viruses with vaccination protection and invivo response to phytohemaglutinin. Genet Sel Evol 22: 457–469.
Gitterle T, degård J, Gjerde B, Rye M, Salte R (2006). Genetic parameters and accuracy of selection for resistance to White Spot Syndrome Virus (WSSV) in Penaeus (Litopenaeus) vannamei using different statistical models. Aquaculture 251: 210–218.
Gjedrem T, Salte R, Gjøen HM (1991). Genetic variation in susceptibility of Atlantic salmon to furunculosis. Aquaculture 97: 1–6.
Gjerde B, Evensen Ø, Bentsen HB, Storset A (2009). Genetic (co)variation of vaccine injuries and innate resistance to furunculosis (Aeromonas salmonicida) and infectious salmon anaemia (ISA) in Atlantic salmon (Salmo salar). Aquaculture 287: 52–58.
Gjøen HM, Refstie T, Ulla O, Gjerde B (1997). Genetic correlations between survival of Atlantic salmon in challenge and field tests. Aquaculture 158: 277–288.
Gudding R, Lillehaug A, Evensen O (1999). Recent developments in fish vaccinology. Vet Immunol Immunopathol 72: 203–212.
Guy DR, Bishop SC, Woolliams JA, Brotherstone S (2009). Genetic parameters for resistance to Infectious Pancreatic Necrosis in pedigreed Atlantic salmon (Salmo salar) post-smolts using a Reduced Animal Model. Aquaculture 290: 229–235.
Håstein T, Gudding R, Evensen (2005). Bacterial vaccines for fish - an update of the current situation worldwide. Dev Biol 121: 55–74.
Houston RD, Haley CS, Hamilton A, Guy DR, Mota-Velasco JC, Gheyas AA et al. (2009). The susceptibility of Atlantic salmon fry to freshwater infectious pancreatic necrosis is largely explained by a major QTL. Heredity 105: 318–327.
Kjøglum S, Henryon M, Aasmundstad T, Korsgaard I (2008). Selective breeding can increase resistance of Atlantic salmon to furunculosis, infectious salmon anaemia and infectious pancreatic necrosis. Aquacult Res 39: 498–505.
Lynch M, Walsh B (1998). Genetics and Analysis of Quantitative Traits. Sinauer Associates: Sunderland.
Madsen PJ, Just J (2007). DMU a package for analysing multivariate mixed models. Department of Genetics and Biotechnology, Research Centre Foulum, University of Aarhus, Faculty Agricultural Sciences (DJF): Tjele.
Moen T, Baranski M, Sonesson A, Kjoglum S (2009). Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics 10: 368.
Nakayama Y, Plisch EH, Sullivan J, Thomas C, Czuprynski CJ, Williams BRG et al. (2010). Role of PKR and Type I IFNs in viral control during primary and secondary infection. PLoS Pathog 6: e1000966.
NOAH 2010. Vaccination of farm animals. Briefing Document No. 22. National Office of Animal Health http://www.noah.co.uk/issues/briefingdoc/bd22pdf.
Nordmo R 1997. Strengths and weakness of different challenge methods. In: Gudding R LA, Midtlyng PJ, Brown F (eds) Fish vaccinology. Dev Biol Stand, Vol. 90 Karger: Basel, Switzerland.
Salte R, Bentsen HB, Moen T, Tripathy S, Bakke TA, Odegard J et al. (2010). Prospects for a genetic management strategy to control Gyrodactylus salaris infection in wild Atlantic salmon (Salmo salar) stocks. Can J Fish Aquat Sci 67: 121–129.
Wetten M, Aasmundstad T, Kjøglum S, Storset A (2007). Genetic analysis of resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar L. Aquaculture 272: 111–117.
Ødegård J, Gitterle T, Madsen P, Meuwissen T, Yazdi M, Gjerde B et al. (2011). Quantitative genetics of Taura syndrome resistance in Pacific white shrimp (Penaeus vannamei): A cure model approach. Genet Sel Evol 43: 14.
Ødegård J, Olesen I, Gjerde B, Klemetsdal G (2007a). Evaluation of statistical models for genetic analysis of challenge-test data on ISA resistance in Atlantic salmon (Salmo salar): prediction of progeny survival. Aquaculture 266: 70–76.
Ødegård J, Olesen I, Gjerde B, Klemetsdal G (2007b). Positive genetic correlation between resistance to bacterial (furunculosis) and viral (infectious salmon anaemia) diseases in farmed Atlantic salmon (Salmo salar). Aquaculture 271: 173–177.
Acknowledgements
This study was funded by grant 179009/S40 from the Research Council of Norway, the Fishery and Aquaculture Industry Research Fund and PHARMAQ AS through the project ‘Does selection for increased disease resistance increase the immune response, the incidence of vaccine injuries and the effect of vaccination?’ SalmoBreed AS provided the fish material and the challenge test data of the three unvaccinated fish groups. The authors are grateful to Celeste Jacq (Nofima) for proof reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Frode Finne-Fridell is a full-time employee at PHARMAQ AS. The remaining authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Drangsholt, T., Gjerde, B., Ødegård, J. et al. Quantitative genetics of disease resistance in vaccinated and unvaccinated Atlantic salmon (Salmo salar L.). Heredity 107, 471–477 (2011). https://doi.org/10.1038/hdy.2011.34
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2011.34
Keywords
This article is cited by
-
Host genetic variation explains reduced protection of commercial vaccines against Piscirickettsia salmonis in Atlantic salmon
Scientific Reports (2020)
-
Moderate heritability of hepatopancreatic parvovirus titre suggests a new option for selection against viral diseases in banana shrimp (Fenneropenaeus merguiensis) and other aquaculture species
Genetics Selection Evolution (2016)
-
Mapping and validation of a major QTL affecting resistance to pancreas disease (salmonid alphavirus) in Atlantic salmon (Salmo salar)
Heredity (2015)
-
Survival, growth and sexual maturation in Atlantic salmon exposed to infectious pancreatic necrosis: a multi-variate mixture model approach
Genetics Selection Evolution (2013)