Abstract
The transfer of genes between Triticum aestivum (hexaploid bread wheat) and T. turgidum (tetraploid durum wheat) holds considerable potential for genetic improvement of both these closely related species. Five different T. aestivum/T. turgidum ssp. durum crosses were investigated using Diversity Arrays Technology (DArT) markers to determine the inheritance of parental A, B and D genome material in subsequent generations derived from these crosses. The proportions of A, B and D chromosomal segments inherited from the hexaploid parent were found to vary significantly among individual crosses. F2 populations retained widely varying quantities of D genome material, ranging from 99% to none. The relative inheritance of bread wheat and durum alleles in the A and B genomes of derived lines also varied among the crosses. Within any one cross, progeny without D chromosomes in general had significantly more A and B genome durum alleles than lines retaining D chromosomes. The ability to select for and manipulate this non-random segregation in bread wheat/durum crosses will assist in efficient backcrossing of selected characters into the recurrent durum or hexaploid genotype of choice. This study illustrates the utility of DArT markers in the study of inter-specific crosses to commercial crop species.
Similar content being viewed by others
Introduction
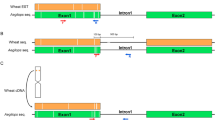
Although cytological investigations commenced in the late nineteenth century, the correct numbers of chromosomes present in hexaploid or bread wheat (Triticum aestivum L; 2n=6x=42) and durum wheat (T. turgidum spp. durum L; 2n=4x=28) were not established until 1918 by Tetsu Sakamura and Karl Sax (Sax, 1918; Kihara, 1982). Soon afterwards the first hexaploid by tetraploid crosses were produced and cytologically examined (Sax, 1922a; Kihara, 1925).
F1 pentaploid hybrids possess chromosomal configurations of 14 bivalents and 7 univalents. The 14 bivalents subsequently behave normally throughout meiosis in F1 plants, whereas the univalent chromosomes split longitudinally at the first division of meiosis (Kihara, 1925). At the second meiotic division the single D chromatids migrate apparently randomly to either pole. Therefore, the number of chromosomes in the gametic nuclei can vary from 14 up to 21. From the F2 generation individuals show varying chromosome counts of between 2n=28 and 2n=42. These numbers stabilize in later generations at either 28 or 42 chromosomes, representing the chromosome numbers of the original parents, and reflect the inherent instability of unpaired chromosomes (Sax, 1922b; Thompson and Hollinghead, 1927; Kihara, 1982). Following this pioneering early research, geneticists have turned their attention elsewhere and only a few recent studies on pentaploid crosses have been published (Gilbert et al., 2000; Wang et al., 2005; Lanning et al., 2008; Eberhard et al., 2010), despite the emergence of new techniques that allow a much more detailed examination of the chromosomal complement and origin.
Wang et al. (2005) analysed the cytological and agronomic characteristics of 55 F2 plants of a durum (female) by hexaploid (male) wheat cross. The F2 progeny ranged in chromosome number from 29 to 42, with an average of 36.5 chromosomes per plant, while the chromosome number distribution frequencies deviated significantly from those reviewed by Kihara (1982). In another study, 10 hexaploid wheat genotypes were crossed with three durum wheat cultivars and the ability of these crosses to produce viable progeny over multiple generations was examined (Lanning et al., 2008). Most of the 182 F5 plants investigated possessed either 28 or 42 chromosomes, with 54 and 36% having the tetraploid and hexaploid complement respectively and only 10% being aneuploids.
As part of a program to transfer resistance to crown rot, caused by the fungus Fusarium pseudograminearum, from a partially resistant hexaploid parent to a susceptible durum background, our group has recently investigated the chromosome composition of the F2 population derived from a T. aestivum/T. turgidum ssp. durum cross (2–49/Bellaroi) using Diversity Arrays Technology (DArT) markers and multicolour fluorescence in situ hybridization (MCFISH) (Eberhard et al., 2010). This study observed that recombination of A and B genome loci from either parent was random in this F2 generation. In addition, a significant number of D genome chromosomes were retained at the F2 stage, with 82 of the 83 F2 progeny investigated possessing some D genome material.
We have now sought to extend this study in several regards. First, we have determined the fate of the retained D genome chromosomes in later generations of the 2–49/Bellaroi population. We have also investigated four other hexaploid/durum crosses using DArT markers to determine whether the degree of D genome retention at F2 and subsequent generations is dependent on either or both of the hexaploid and tetraploid parents. Finally, we have compared the relative incorporation of the A and B genomes of each parent in the progeny to determine whether the random segregation of parental materials, observed previously in one population (Eberhard et al., 2010), is typical of hexaploid/durum crosses.
Materials and methods
Plant material
The five different hexaploid/durum (T. aestivum/T. turgidum spp. durum) crosses examined in this study were produced under the National Durum Breeding Program based at the Tamworth Agricultural Institute, Industry and Investment New South Wales, Tamworth, Australia (Table 1). For each cross, a hexaploid wheat line with partial crown rot resistance was used as the maternal parent. Subsequent generations of the 2–49/950329 cross were produced to the F6 stage and of the 2–49/Bellaroi and CPI133814/Bellaroi crosses to the F7 stage. F2 generations from two Sunco/durum crosses were also included in this study (Table 1).
Seeds were germinated in a 24-well tissue culture plate at 24 °C in the dark. Leaf material (about 50 mg) was sampled from 1-week-old seedlings. Differences in sample numbers (Table 1) are the result of seed availability and differences in germination rates. DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega Corporation, Sydney, Australia). The DNA was quantified using an Implen Nanophotometer, Integrated Sciences, Sydney, Australia.
Diversity Arrays Technology
The DNA of the lines listed in Table 1 was sent to Triticarte P/L, Canberra (http://www.triticarte.com.au/default.html) for DArT analysis using a hexaploid wheat array (Wenzl et al., 2004). Only markers with known chromosome location were included in the analyses (Akbari et al., 2006). A Q-value for each marker, which is an estimate of the marker quality, was supplied by Triticarte P/L. The Q-value reflects how well the two phases (that is, present=1 versus absent=0) of the marker are separated in a sample set. In this study, only markers with a Q-value > 77 were used in the analyses, as this is the suggested threshold value for reliability (Akbari et al., 2006). Goodness-of-fit for each DArT marker was tested by means of a chi-square analysis. For the 2–49/Bellaroi F2 population a genetic linkage map was produced. MapManager QTXb20 (Manly et al., 2001) was employed to partition DArT markers into linkage groups and RECORD (Van Os et al., 2005) was used to order markers within linkage groups. The Kosambi function was applied to calculate map distances. Map figures were produced with MapChart version 2.1 (Voorrips, 2002).
Data analysis
Univariate analysis of variance to test for differences between means was conducted using SPSS version 18. Graphs were constructed in Microsoft Excel 2007.
Results
The number of polymorphic DArT markers used in the analyses of the different populations ranged from 162 to 458 (Table 1 and Supplementary Table 1). A hexaploid wheat array was used as the marker source to be able to determine the D genome constitution of the progeny. Between 12 and 37% of the polymorphic markers in this array hybridized with the durum DNA, depending on the population (Table 1). A genetic map of the A and B genomes of the 2–49/Bellaroi F2 population is presented as Supplementary Figure 1.
D genome content
Significant differences were observed in the D genome content of the different populations (Table 2). In all, 99 and 92% of the lines in the F2 (Eberhard et al., 2010) and F3 2–49/Bellaroi populations, respectively, inherited at least one D genome chromosome. Of these lines, 52% (F2) and 61% (F3) of the independent samples possessed all seven D genome chromosomes (Figure 1). A relatively low frequency of partial chromosomes was also observed in these populations (Table 2). The number of lines with at least one D chromosome in the 2–49/Bellaroi F7 population decreased to 31% (Table 2), of which all but one (missing 5D) possessed all seven D chromosomes (Figure 1). No partial chromosomes were observed at the F7 generation. Similar results were obtained with the F6 generation of the 2–49/950329 population (Table 2; Figure 1). Three partial 5D chromosomes were present in this population. In the Sunco/Bellaroi F2 population 88% of the lines had at least one D chromosome. In contrast to the 2–49/Bellaroi F2 population, only 6% of the Sunco/Bellaroi lines possessed at least one copy of all seven D chromosomes while the mode (29%) was five D chromosomes (Figure 1). None of the lines of the Sunco/230604 F2 or the F7 CPI133814/Bellaroi populations possessed any D chromosomes, whereas only two lines of the F3 CPI133814/Bellaroi population retained D genome material, which in both cases was a single chromosome, 7D (Table 2). The group (1–7) to which the remnant D chromosomes belonged appeared to be random in all populations that retained significant quantities of D chromatin.
The F2 and F3 2–49/Bellaroi populations possessed the highest average number of D chromosomes per line, 5.3 and 5.9, respectively (Table 2). This number decreased to 2.1 in the F7 population of this cross, a similar level to that observed in the F6 generation of the 2–49/950329 cross.
Segregation distortion
Chromosomes were considered to be associated with skewed segregation of parental alleles if four or more markers on a chromosome showed segregation distortion. Normal segregation (P<0.01) was observed with most of the A and B genome markers of the 2–49/Bellaroi, 2–49/950329 and Sunco/Bellaroi populations (Table 3). In contrast, 58% and up to 36% of the markers of the Sunco/230604 and CPI133814/Bellaroi populations, respectively, showed segregation distortion. Distortion in these populations was always in favour of the durum parent, with the exception of the markers on chromosome 6A in the CPI133814/Bellaroi F3 population. The identity of chromosomes involved in segregation distortion appeared to be random except for markers on chromosome 5B. Markers in four out of the five populations examined showed distortion in the same region on chromosome 5B in favour of the durum parent (Table 3 and Supplementary Figure 1).
Percentage of A and B durum alleles present in each population
All plants possessed alleles from both parents in their A and B genome, indicating that there were no maternal self-pollinations when the parental crosses were made. The percentage of durum parental alleles present in successive generations of the 2–49/Bellaroi population increased from a mean of 34% for the F3 plants to 52% in the F7 generation (Figure 2). An average of 52% durum alleles was also observed in the 2–49/950329 F6 population (Figure 2). The Sunco/Bellaroi population possessed a similar proportion of durum alleles as the early generations of the 2–49/Bellaroi cross (38%; Figure 2). In contrast, the mean proportion of durum alleles in the Sunco/230604 and both CPI133814/Bellaroi generations was much higher, at 76% and 61–68%, respectively (Figure 2).
Box plot distributions indicating the percentage of A and B genome durum alleles present in each population. The range is indicated by vertical lines, the 25 and 75 percentiles by the horizontal box boundaries, and the median by the solid horizontal line. Boxes sharing the same letter are not significantly different (P>0.05).
Discussion
Studies determining the chromosome content of progeny of pentaploid crosses were conducted as early as the 1920s (Sax, 1922b; Thompson and Hollinghead, 1927; Kihara, 1982). Although these authors could estimate the number of bivalent and univalent chromosomes using traditional cytological methods at meiosis, they could not routinely determine the identity of the chromosomes, nor could they easily detect intergenomic translocations. Recently developed molecular markers based on DArT analysis now readily permit these investigations. However, due to the dominant nature of DArT arrays, supplementary cytological methods are still useful for distinguishing between single and multiple copies of D genome chromosomes (Eberhard et al., 2010).
The total number of lines having at least one D genome chromosome was similar in the F2 and F3 2–49/Bellaroi and F2 Sunco/Bellaroi populations. However, the number of unique chromosomes present in the lines of these populations differed (Figure 1). More than 50% of the progeny of the 2–49/durum populations had at least one copy of all seven D chromosomes, whereas the most frequent number of unique D chromosomes present in the progeny of the Sunco/230604 cross was five. In the F6 and F7 generations of the 2–49/950329 and 2–49/Bellaroi crosses, 70% of the lines were tetraploids and the rest, with the exception of three lines, all carried at least one copy of all seven D chromosomes. This result is similar to those observed by Lanning et al. (2008). They found that more than 50% of the F5 progeny produced from 10 different hexaploid/durum crosses were tetraploids and 10% were aneuploid.
The average number of unique D genome chromosomes present in crosses to the hexaploid 2–49 parent was higher than for all other parental combinations. Whereas the Sunco/Bellaroi population retained half the number of D chromosomes compared with lines from crosses involving 2–49, progeny from the Sunco/230604 and CPI133814/Bellaroi crosses had lost all or nearly all D genome material by the F2 and F3 generations, respectively. These results imply that particular parental combinations have an important role in determining the retention of the D genome in subsequent generations derived from pentaploid crosses. Furthermore, the identity of both the hexaploid and tetraploid parent will influence the degree of D genome retention.
Using DArT markers, which produce a whole genome profile, this study has identified a number of partial chromosomes in the early generation progeny of these crosses. Comparisons between DArT marker data and MCFISH-based cytological analyses using the same lines have indicated that these partial chromosomes can be readily identified as either telocentric chromosomes or translocations (Eberhard et al., 2010). Only one partial chromosome was detected in the F6 and F7 materials examined, perhaps indicating that this represents a stable translocation event, considering that independent chromosome fragments are frequently lost during meiosis.
Minimal segregation distortion in the A and B genomes was observed with the DArT markers in three of the five crosses investigated. In contrast, 58% of the markers of the Sunco/230604 and up to 36% of the CPI133814/Bellaroi populations were distorted mostly in favour of the durum parent. A segregation distortion of up to 23% has been observed in other studies of hexaploid/hexaploid crosses (Cadalen et al., 1997; Semagn et al., 2006; Mantovani et al., 2008; Peleg et al., 2008). These segregation outcomes are likely to arise from particular parental interactions and provide opportunities for manipulating crosses so that the progeny show higher levels of the more desirable parental background. Markers in four of the five crosses from which these populations were derived indicated segregation distortion on chromosome 5B in favour of the durum parent. Segregation distortion has previously been detected with loci on chromosome 5B in tetraploid/tetraploid crosses (Kumar et al., 2007). Kumar et al. (2007) found that different distorter or meiotic drive elements exist among different genotypes and that distortion factors occur at different locations along chromosome 5B and also differ in underlying mechanisms.
An increase of 12% in durum alleles was observed from the F3 to the directly derived F7 generation of the 2–49/Bellaroi cross. This is likely to be due to the effects of several rounds of selection towards the durum head type in this population. Particular combinations of durum-derived chromosome segments may also be more compatible and fertile, leading to higher survival rates. Interestingly, the populations with the least number of D chromosomes possessed the highest percentage of durum alleles in their A and B genomes.
A better understanding of the mechanisms that control both D genome retention and the proportion of A and B genomes derived from the durum parent will assist durum breeders in selecting parental combinations that will favour inheritance of durum alleles in the progeny. At the same time, reliable phenotypic or molecular markers will permit the transfer of desired traits from the hexaploid parent into the durum parental background. This ability to manipulate non-random segregation of the two genomic sources will require fewer generations to transfer selected characters into the recurrent parent of choice, using an approach based on accelerated marker-assisted selection of both background and introgressed traits.
In summary, we have investigated five different hexaploids by durum crosses at a variety of subsequent generations and have identified differences in the amount of D genome material retained and the percentage of the durum parent present in the progeny. This implies that crosses such as Sunco/230604 and CPI133814/Bellaroi may be more useful when the aim is to transfer genes from the A and B genome of hexaploid wheat into durum wheats, as the progeny do not retain D chromosomes and have a higher percentage of the durum parent in their background. Conversely, crosses such as 2–49/Bellaroi and 2–49/950329, which retain a high number of D chromosomes and inherit a lower proportion of the durum background, may be more useful in breeding strategies using durum wheat as a trait source for hexaploid wheat improvement. Finally, the findings of this study will have application not just to international cereal breeding but also across other crop species where interspecific crosses to wild relatives are employed to introgress novel characters.
References
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S et al. (2006). Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113: 1409–1420.
Cadalen T, Boeuf C, Bernard S, Bernard M (1997). An interval molecular marker map in Triticum aestivum LEm. Thell. and comparison with a map from a wide cross. Theor Appl Genet 94: 367–377.
Eberhard F, Zhang P, Lehmensiek A, Hare R, Simpfendorfer S, Sutherland MW (2010). Chromosome composition of an F2 Triticum aestivum x Tturgidum spp. durum cross analysed by DArT markers and MCFISH. Crop Pasture Sci 61: 619–624.
Gilbert J, Procunier JD, Aung T (2000). Influence of the D genome in conferring resistance to fusarium head blight in spring wheat. Euphytica 114: 181–186.
Kihara H (1925). Weitere Untersuchungen über die pentaploiden Triticum-Bastarde. I. Jpn J Bot 2: 299–305.
Kihara H (1982). Wheat Studies-Retrospect and Prospects. Kodansha Ltd: Tokyo.
Kumar S, Gill BS, Faris JD (2007). Identification and characterization of segregation distortion loci along chromosome 5B in tetraploid wheat. Mol Genet Genomics 278: 187–196.
Lanning SP, Blake NK, Sherman JD, Talbert LE (2008). Variable production of tetraploid and hexaploid progeny lines from spring wheat by durum wheat crosses. Crop Sci 48: 199–202.
Manly KF, Cudmore JRH, Meer JM (2001). Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12: 930–932.
Mantovani P, Maccaferri M, Sanguineti MC, Tuberosa R, Catizone I, Wenzl P et al. (2008). An integrated DArT-SSR linkage map of durum wheat. Mol Breeding 22: 629–648.
Peleg Z, Saranga Y, Suprunova T, Ronin Y, Roder MS, Kilian A et al. (2008). High-density genetic map of durum wheat x wild emmer wheat based on SSR and DArT markers. Theor Appl Genet 117: 103–115.
Sax K (1918). The behaviour of the chromosomes in fertilization. Genetics 3: 309–327.
Sax K (1922a). Sterility in wheat hybrids. III. Endosperm development and F2 sterility. Genetics 7: 554–559.
Sax K (1922b). Sterility in wheat hybrids. II. Chromosome behaviour in partially sterile hybrids. Genetics 7: 513–552.
Semagn K, Bjornstad A, Skinnes H, Maroy AG, Tarkegne Y, William M (2006). Distribution of DArT, AFLP, and SSR markers in a genetic linkage map of a doubled-haploid hexaploid wheat population. Genome 49: 545–555.
Thompson WP, Hollinghead L (1927). Preponderance of dicoccum-like characters and chromosome numbers in hybrids between Triticum dicoccum and T. vulgare. J Genet 17: 283–307.
Van Os H, Stam P, Visser RGF, Van Eck H (2005). RECORD: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet 112: 30–40.
Voorrips RE (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93: 77–78.
Wang H-Y, Liu D-C, Yan Z-H, Wei Y-M, Zheng Y-L (2005). Cytological characteristics of F2 hybrids between Triticum aestivum L and T. durum Desf. with reference to wheat breeding. J Appl Genet 46: 365–369.
Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A et al. (2004). Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci USA 101: 9915–9920.
Acknowledgements
This work was partially funded by the Grains Research and Development Corporation, Australia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Supplementary information
Rights and permissions
About this article
Cite this article
Martin, A., Simpfendorfer, S., Hare, R. et al. Retention of D genome chromosomes in pentaploid wheat crosses. Heredity 107, 315–319 (2011). https://doi.org/10.1038/hdy.2011.17
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2011.17
Keywords
This article is cited by
-
Asymmetric gene expression in grain development of reciprocal crosses between tetraploid and hexaploid wheats
Communications Biology (2022)
-
Association between presence of Triticum timopheevii introgression and D-genome retention in hexaploid/tetraploid wheat crosses
Molecular Breeding (2018)
-
Genome inheritance in populations derived from hexaploid/tetraploid and tetraploid/hexaploid wheat crosses
Molecular Breeding (2017)
-
Evaluating interspecific wheat hybrids based on heat and drought stress tolerance
Journal of Crop Science and Biotechnology (2016)
-
Microsatellite mapping of the gene for sham ramification in spikelets derived from a hexaploid wheat (Triticum spp.) accession 171ACS
Genetic Resources and Crop Evolution (2015)