Abstract
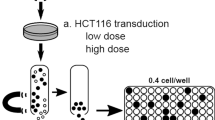
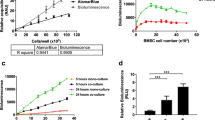
RNA interference (RNAi) is a sequence-specific gene silencing mechanism with therapeutic potential against many human pathogens. To obtain a durable therapeutic effect, stable transduction of target cells with for instance a lentiviral vector that expresses a short hairpin (shRNA) inducer of the RNAi pathway is necessary. Apart from the intended therapeutic effect, this treatment can induce negative effects on cell proliferation via off-target effects. A careful evaluation of the transduced cells is required to develop a safe gene therapy approach. Stably transduced cells are usually selected by expression of the enhanced green fluorescent protein (GFP) marker. In this study we show that the mixed transduction culture, containing both transduced GFP+ and untransduced GFP− cells, can simply be passaged to score the GFP+/GFP− ratio by longitudinal flow cytometric analysis as a measure of the negative impact of the RNAi treatment on the cellular proliferation rate. We show that this assay is sensitive, easy to use and internally controlled for assessing subtle effects on cell proliferation of lentiviral transduction and transgene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC . Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391: 806–811.
Liu YP, Haasnoot J, ter BO, Berkhout B, Konstantinova P . Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res 2008; 36: 2811–2824.
Zhiqiang W, Yaowu Y, Fan Y, Jian Y, Yongfeng H, Lina Z et al. Effective siRNAs inhibit the replication of novel influenza A (H1N1) virus. Antiviral Res 2010; 85: 559–561.
Pan Q, Henry SD, Metselaar HJ, Scholte B, Kwekkeboom J, Tilanus HW et al. Combined antiviral activity of interferon-alpha and RNA interference directed against hepatitis C without affecting vector delivery and gene silencing. J Mol Med 2009; 87: 713–722.
Eckstein A, Grossl T, Geisler A, Wang X, Pinkert S, Pozzuto T et al. Inhibition of adenovirus infections by siRNA-mediated silencing of early and late adenoviral gene functions. Antiviral Res 2010; 88: 86–94.
Liu YP, von Eije KJ, Schopman NC, Westerink JT, ter BO, Haasnoot J et al. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol Ther 2009; 17: 1712–1723.
Eekels JJ, Geerts D, Jeeninga RE, Berkhout B . Long-term inhibition of HIV-1 replication with RNA interference against cellular co-factors. Antiviral Res 2011; 89: 43–53.
Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 2003; 21: 635–637.
Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006; 441: 537–541.
Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS . Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 2009; 27: 549–555.
Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I . Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol 2005; 23: 457–462.
Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med 2005; 11: 263–270.
Sioud M . Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol 2005; 348: 1079–1090.
He J, Yang Q, Chang LJ . Dynamic DNA methylation and histone modifications contribute to lentiviral transgene silencing in murine embryonic carcinoma cells. J Virol 2005; 79: 13497–13508.
Ellis J . Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther 2005; 16: 1241–1246.
ter Brake O, Konstantinova P, Ceylan M, Berkhout B . Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther 2006; 14: 883–892.
Mosmann T . Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983; 65: 55–63.
Crouch SP, Kozlowski R, Slater KJ, Fletcher J . The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J Immunol Methods 1993; 160: 81–88.
Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM . Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods 2006; 3: 715–719.
Seppen J, Rijnberg M, Cooreman MP, Oude Elferink RP . Lentiviral vectors for efficient transduction of isolated primary quiescent hepatocytes. J Hepatol 2002; 36: 459–465.
Ruijter JM, Thygesen HH, Schoneveld OJ, Das AT, Berkhout B, Lamers WH . Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology 2006; 3: 2.
Acknowledgements
This research was supported by the Dutch AIDS fund (grant 2006006 and 2007028). We thank Berend Hooibrink for FACS sorting, Stephan Heynen for CA-p24 enzyme-linked immunosorbent assay experiments and Renée van der Sluis and Dave Speijer for useful discussions. We also thank the Belgian Federal Government for financial support through the Inter-University Attraction Pole grant P6/41.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Gene Therapy website
Supplementary information
Rights and permissions
About this article
Cite this article
Eekels, J., Pasternak, A., Schut, A. et al. A competitive cell growth assay for the detection of subtle effects of gene transduction on cell proliferation. Gene Ther 19, 1058–1064 (2012). https://doi.org/10.1038/gt.2011.191
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2011.191
Keywords
This article is cited by
-
A biallelic multiple nucleotide length polymorphism explains functional causality at 5p15.33 prostate cancer risk locus
Nature Communications (2023)
-
GLUT5 (SLC2A5) enables fructose-mediated proliferation independent of ketohexokinase
Cancer & Metabolism (2021)
-
The miR-17/92 cluster is involved in the molecular etiology of the SCLL syndrome driven by the BCR-FGFR1 chimeric kinase
Oncogene (2018)
-
Silencing of HIV-1 by AgoshRNA molecules
Gene Therapy (2017)
-
MAP3K11 is a tumor suppressor targeted by the oncomiR miR-125b in early B cells
Cell Death & Differentiation (2016)