Abstract
Background:
Chromosome 15q13.3 represents a hotspot for genomic rearrangements due to repetitive sequences mediating nonallelic homologous recombination. Deletions of 15q13.3 have been identified in the context of multiple neurological and psychiatric disorders, but a prospective clinical and behavioral assessment of affected individuals has not yet been reported.
Methods:
Eighteen subjects with 15q13.3 microdeletion underwent a series of behavioral assessments, along with clinical history and physical examination, to comprehensively define their behavioral phenotypes.
Results:
Cognitive deficits are the most prevalent feature in 15q13.3 deletion syndrome, with an average nonverbal IQ of 60 among the patients studied. Autism spectrum disorder was highly penetrant, with 31% of patients meeting clinical criteria and exceeding cutoff scores on both ADOS-2 and ADI-R. Affected individuals exhibited a complex pattern of behavioral abnormalities, most notably hyperactivity, attention problems, withdrawal, and externalizing symptoms, as well as impairments in functional communication, leadership, adaptive skills, and activities of daily living.
Conclusions:
The 15q13.3 deletion syndrome encompasses a heterogeneous behavioral phenotype that poses a major challenge to parents, caregivers, and treating providers. Further work to more clearly delineate genotype–phenotype relationships in 15q13.3 deletions will be important for anticipatory guidance and development of targeted therapies.
Genet Med 18 11, 1111–1118.
Similar content being viewed by others
Introduction
The long arm of chromosome 15 is particularly notable for its role in human-specific behavior and neurodevelopmental/psychiatric disorders. The unique genomic architecture of the 15q11–q13 segment, which contains six low-copy repeat elements, results in frequent duplications and deletions;1 these have been associated with a broad spectrum of behavioral phenotypes in humans.2 The unique dosage-dependent nature of these phenotypes, their heterogeneous manifestations, and the frequency with which structural rearrangements occur in this region make it of great interest with respect to understanding the genetic contribution to human-specific behavior as well as to neurobehavioral disorders such as autism spectrum disorder (ASD) and schizophrenia.
In particular, the 15q13.3 locus has gained attention because of its role in human-specific behaviors and neurobehavioral phenotypes. Although the pathogenicity of duplications of that segment is uncertain, deletions involving breakpoint (BP) region BP4-BP5 of 15q13.3 appear to cause neurobehavioral disorders with relatively high penetrance.3,4,5 Overall, it is estimated that the prevalence of 15q13.3 microdeletions is 1 in 40,000 among the general population and as high as 1 in 450–600 for samples submitted for clinical CMA,6,7,8,9 necessitating a thorough understanding of the clinical phenotype. Many reports of patients with 15q13.3 microdeletions have been published, but systematic characterization of the behavioral phenotypes associated with deletions in this region has not been undertaken, despite neurodevelopmental disorders being consistently reported. For instance, a recent meta-analysis of more than 200 adult and pediatric cases with deletions in the 15q13.3 region suggested that behavioral manifestations are very common, most notably developmental delay/intellectual disability, epilepsy/seizures, and speech problems. Congenital anomalies were found in only six patients, with congenital heart disease being most frequent.10 The first reports of patients with 15q13.3 deletion syndrome described individuals with the predominate phenotype of intellectual disability and seizures/epilepsy, but there was also a notable proportion of individuals with ASD and other neuropsychiatric problems, although the frequencies of these psychiatric problems varied widely between studies.4,11,12,13,14,15 Moreover, enrichment of 15q13.3 deletions among patients with schizophrenia has been reported.16,17 Therefore, it has become clear that the major phenotypes of 15q13.3 deletion syndrome are neurobehavioral; however, a systematic assessment of the prevalence and characteristics of the 15q13.3 microdeletion syndrome phenotype is still lacking, as is an understanding of the genotype–phenotype relationship underlying these behavioral manifestations.
The low-copy repeat elements in the 15q11–q13 region cluster into six breakpoints (termed BP1–BP6), three of which are located in the 15q13 locus (BP3–5; Figure 1 ). The most commonly reported rearrangements in this locus are deletions encompassing only BP4–BP5, but larger deletions of BP3–BP5 have also been reported.18,19,20 There are six RefSeq genes in the BP4-BP5 region (MTMR15, MTMR10, TRPM1, KLF13, OTUD7A, and CHRNA7) and several noncoding RNAs and other poorly characterized elements ( Figure 1 ). Although the exact contribution of the genes in this region to these described phenotypes remains unclear, we previously demonstrated that smaller deletions encompassing only the CHRNA7 gene, encoding an alpha-7 nicotinic receptor with high expression in the brain, have phenotypic consequences similar to those of the larger BP4–BP5 deletion, including intellectual disability, seizures, aggressive behavior, and attention-deficit hyperactivity disorder (ADHD), suggesting CHRNA7 as a potential candidate gene for the observed phenotypes of the 15q13.3 microdeletion syndrome.21 Other work also supports the notion that CHRNA7 probably plays a central role in the phenotype.3,22 However, it is important to note that other genes in the region have been posited as contributing to the neuropsychiatric phenotype, such as rare nonsynonymous variants in FAN1 identified in cases of schizophrenia and ASD.23 Although CHRNA7 and other candidates have shown an intriguing relationship to neurobehavioral phenotypes, there remain insufficient data showing clear genotype–phenotype correlations within the 15q13.1–q13.3 region.10
The evidence suggests that the 15q13.3 region makes unique contributions to human neurodevelopment and behavior; therefore, a thorough understanding of rearrangements in this region and their corresponding phenotypes is important. The goal of this work was to more comprehensively define the medical, neuropsychological, and behavioral phenotypes of individuals with 15q13.3 microdeletions. To do this, we employed a genotype-to-phenotype approach whereby we identified individuals aged 12–18 years who had been found by chromosomal microarray (CMA) to have 15q13.3 microdeletions mainly encompassing BP4–BP5; we then performed a systematic assessment of their medical and behavioral phenotypes. Our results represent the most comprehensive, systematic assessment of a group of patients with 15q13.3 microdeletions to date. The complex behavioral phenotype we describe highlights some of the challenges affected individuals and their families face on a daily basis. At the same time, this study may help physicians and genetic counselors discuss the implications of 15q13.3 deletions and provide better anticipatory guidance. Finally, the complex nature of the clinical manifestations associated with this copy-number variant (CNV) should be taken into consideration when planning clinical trials for affected individuals.
Materials and Methods
This study was approved by the institutional review board at Baylor College of Medicine. Patients with 15q13.3 microdeletions spanning at least BP4–BP5 were identified retrospectively by review of CMA results performed in the Baylor Medical Genetics Laboratory or Signature Genomics Laboratories (prior to its closure in June 2014), and their referring providers were contacted. A total of 18 patients (aged 12–18 years; Tables 1 and 2 ) agreed to participate in this study and attended a 2-day medical and behavioral comprehensive assessment at Texas Children’s Hospital. The study involved a single dose of a pharmaceutical compound on day 2 of the assessment. The results of that assessment will be reported separately. Patients 12 and 16 are biological brothers. The remaining 16 study participants were unrelated to one another. The total number of 15q13.3 microdeletions spanning at least BP4–BP5 was 98 (of a total of 57,711 cases submitted for chromosome microarray analysis) at the Baylor Medical Genetics Laboratory and 114 (of 46,145) at Signature Genomic Laboratories.
Each patient had previously had CMA ordered for various indications ( Table 2 ). Secondary CNVs were detected in 5 of the 18 study participants (see Supplementary Table S1 online) and reported as findings of uncertain clinical significance on the respective clinical CMA reports. Because CMA studies of the study participants were performed as clinical tests at different time points, resolution and interpretation of findings were subject to variation and may have affected the reporting of secondary CNVs.
To confirm the 15q13.3 genotypes of all probands in this study, a peripheral venous blood sample was obtained and genomic DNA was isolated and subjected to multiplex ligation-dependent probe amplification (MLPA). Parents of each patient who were available and willing to participate were also tested with multiplex ligation-dependent probe amplification (MLPA) for CHRNA7 copy number to determine the inheritance pattern.
MLPA reactions were performed according to the manufacturer’s instructions (MRC-Holland, Amsterdam, The Netherlands). A total of 12 pairs of synthetic probes were designed to detect CHRNA7 exon 2, exon 4, exon 5, and exon 10, KLF13 exon 2, OTUD7A exon 9, TRPM1 exon 1, MTMR10 exon 14, MTMR15 exon 2 and exon 14, TJP1 exon 12, NDNL2 exon 1, and APBA2 exon 14 (see Supplementary Table S2 online). Ligation quality (D-fragment) and DNA quality (Q-fragments) were assessed for the DNA fragment analysis of each reaction. Calculated dosage quotient (DQ) reflects the amount of fluorescence signal detected within the DNA fragment analysis. The ratio of the peak height of the synthetic or reference probe over the average of 14 reference probes of a sample was determined. The value of this ratio was then divided by the average of the corresponding probe of all samples. Reference DQ values were provided by MRC-Holland to identify potential deletions or duplications. A heterozygous deletion is defined as a DQ between 0.35 and 0.65, and a heterozygous duplication is defined as DQ between 1.35 and 1.55. Normal copy-number status is a DQ reference value between 0.85 and 1.15.
Probands were evaluated by a team of providers at Texas Children’s Hospital. Clinical history and physical examination were obtained by a single provider (C.P.S.). Cognitive testing and behavioral testing were administered by collaborating psychologists (R.P.K. and L.N.B.) and involved the Differential Ability Scales–II; the Autism Diagnostic Interview–Revised (ADI-R); the Autism Diagnostic Observation Schedule, Second Edition (ADOS-2); the Adaptive Behavior Assessment System, Second Edition (ABAS-II); and the Behavioral Assessment for Children, Second Edition (BASC-2). Examiners administering the ADI-R and ADOS-2 were research reliable on both instruments. Patient characteristics, IQ measures, and behavioral scales were summarized using frequency with percentage, mean with standard deviation, and median with 25th and 75th percentiles. A one-sample t-test was used to compare the sample mean with the mean of the normative score.
Results
Genetics
The patients recruited into this study all had deletions encompassing the BP4–BP5 regions of 15q13.3, except for three patients (patients 4, 12, and 16), whose deletions were larger (BP3–BP5). For six of the patients, we were unable to obtain genetic material from either parent to assess inheritance patterns. For five individuals, only one parent was available (four mothers and one father), who was found not to carry the deletion. Six individuals had inherited the 15q13.3 deletion from their mothers, five of whom had a history of neuropsychiatric disease themselves, and one did not report any cognitive or behavioral problems (but was not formally tested). One study participant was found to have a de novo deletion ( Table 1 ). This distribution is similar to those in previously published reports.21 Five of the 18 patients (28%) had a second CNV of uncertain clinical significance discovered outside of the 15q13 region (see Supplementary Table S1 online).
Clinical characteristics
Of the 18 probands who underwent comprehensive assessment, 14 were male and 4 were female, with an average age of 14 years (SD 1.7) ( Table 2 ). Average height, weight, and fronto-occipital circumference of the patients studied were within normal limits. Clinically, the most common medical comorbidity was epilepsy/seizures (n = 5, 28%), followed by hypotonia as an infant (n = 3, 17%). Two patients had a history of undescended testicles, two patients had cardiac malformations, and one patient each had cardiac arrhythmia, cleft palate, and ectodermal dysplasia. More than half of the patients (n = 10) were originally referred for CMA testing for intellectual disability/developmental delay ( Table 2 ). Other reasons for initial referral included behavioral problems (4), autism (2), and bipolar disorder (1). Four patients reported concurrent ADHD; one patient had only hyperactivity without attention-deficit disorder. Developmentally, on average, the patients sat at 10 months, crawled at 12 months, walked at 19 months, spoke their first word at 26 months, and formed two-word sentences at 44 months (see Supplementary Table S3 online). No consistent dysmorphic facial features were noted among the patients studied ( Figure 2 ).
Behavioral phenotype
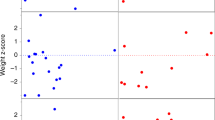
Intellectual ability was assessed using the Differential Ability Scales–II. Figure 3 shows histograms of the ratio IQ measure reported here (ratio IQ = (mental age/chronological age) × 100), with the curve of the normative scores using a normal distribution with a mean of 100 and standard deviation of 15. The average verbal ratio IQ was 64.3 (95% CI: 52.8, 75.8) and was statistically different from the normative average of 100 (P < 0.001). The average nonverbal reasoning ratio IQ was 60.1 (95% CI: 49.6, 70.6) and was statistically different from the normative average (P < 0.001). The full-scale reasoning IQ average was also different from the normative (P < 0.001), with an average of 60.9 (95% CI: 50.3, 71.6). The verbal and nonverbal subcomponents of the IQ test were not significantly different from each other, although this may be due to the small sample size.
Histograms of the composite scores with a curve of the normative scores using a normal distribution with mean of 100 and standard deviation of 15 for full-scale reasoning IQ (FSRIQ), verbal (VRIQ) and nonverbal reasoning IQ (NVRIQ) scores, and the Behavioral Assessment for Children, Second Edition (BASC) behavioral assessment, with a normative mean of 50 and standard deviation of 10. DAS, Differential Ability Scale.
Behavioral phenotype was assessed using the BASC-2 and the ABAS-II tests. Figure 3 shows a histogram of the composite BASC-2 scores, with the curve of the normative scores using a normal distribution with a mean of 50 and standard deviation of 10. The average Behavioral Symptoms Index T score was 67.1 (95% CI: 61.2, 72.9), which is statistically different from the normative average (P < 0.001). Subcomponents of the BASC-2 were also significantly different (see Supplementary Table S4 online), including the average Externalizing T score of 63.6 (95% CI: 56.9, 70.3; P = 0.0006), the average Internalizing T score of 62.9 (95% CI: 52.8, 72.9; P = 0.0154), and the average Adaptive T score of 32.4 (95% CI: 30.5, 34.2, P < 0.001). The average General Adaptive Composite Score was 64 (95% CI: 53.9, 74.1), which is statistically different from the normative average (P < 0.001). The other three composite scores (from ABAS-II) were also statistically different from the normative average (all P < 0.001, see Supplementary Table S5 online), including the average Conceptual Composite Score (66.9; 95% CI: 56.3, 77.5), the average Social Composite Score (69.4; 95% CI: 58.1, 80.8), and the average Practical Composite Score (59.9; 95% CI: 47.4, 72.3). We also assessed for specific behaviors altered in patients using the BASC-2. The behaviors most remarkably altered in this cohort were hyperactivity (87.7 percentile, SD 12.6), attention problems (84.6 percentile, SD 11.3), social withdrawal (83.6 percentile, SD 22), externalizing problems (82.3 percentile, SD 18.3), and deficits in functional communication (8.9 percentile, SD 8.8), leadership (7.9 percentile, SD 7.7), activities of daily living (5.8 percentile, SD 6.4), and adaptive skills (5.0 percentile, SD 2.9).
Patients were assessed for ASD using two gold-standard assessments, one by direct observation (ADOS-2) and one by parent report (ADI-R). In addition, the clinical impression of an expert clinical psychologist was given. Overall, 31% of patients met criteria for ASD by both ADOS-2 and ADI-R (39% of patients by ADOS-2 alone and 50% by ADI-R alone), and 44% of patients were given a diagnosis of ASD by clinical impression. The content areas of ADI-R are summarized in Supplementary Table S5 online, which describes them both continuously and according to cutoff values.
Discussion
We show that the 15q13.3 microdeletion syndrome encompasses a complex and heterogeneous pattern of behavioral abnormalities, with cognitive deficits being most prevalent. Behavioral abnormalities, notably ASD, were also very common, and many individuals also had seizures/epilepsy. No distinct dysmorphic characteristics appear to define this phenotype in our study cohort; nor are verbal abilities particularly more affected than general cognitive function.
These results provide important insights into the complex behavioral phenotype of 15q13.3 microdeletion syndrome. Variable expressivity and phenotypic heterogeneity are observed in most, if not all, copy-number changes predisposing to neuropsychiatric disease, and that holds true for 15q13.3 microdeletion syndrome as well. For instance, although the average full-scale IQ was 60 overall, the range was quite large (25–88), suggesting that the phenotypic expressions are not only the result of the specific CNV itself but also subject to modification and/or interaction with other factors affecting brain development. Potential modifiers could include other genes or gene–environment interactions. Second CNVs outside of the 15q13.3 locus represent one example of a modifier, and 28% of the participants in this study were found to carry a second CNV, which was interpreted as a variant of uncertain clinical significance and identified by clinical chromosome microarray analysis. Future work to identify and characterize additional genetic and nongenetic modifiers will be necessary.
Among the 18 study participants, only 4 were female. One may speculate that female individuals with 15q13.3 microdeletion are less likely to present for clinical assessment due to milder clinical consequences, as suggested by a “female protective model” for neurodevelopmental disorders in general.22 This is supported by the increased maternal carrier incidence in this study (six maternal carriers versus no paternal carrier identified).
The prevalence of ASD in this syndrome was particularly high, with 31% of probands in this study meeting criteria as defined by cutoff scores for both the ADOS-2 and ADI-R. This rate is higher than that reported in similar microdeletion syndromes with prominent neurobehavioral phenotypes, such as 16p11.2 deletion, despite resembling strategies for both enrollment and assessment in the respective studies.24 This finding is important both for providing anticipatory guidance to families/caregivers of the affected children and for furthering our understanding of the complex genetic contributions to the broad autism phenotype. Subtle differences in patient recruitment may account for some of those differences. The participants of this study were invited to participate in a study that included a brief clinical trial of a pharmaceutical compound, whereas the 16p11.2 study was purely diagnostic. Also, it should be noted that the age range of participants in this study was quite narrow (12–18 years).
It is notable that a high proportion of the probands (one-third) was adopted. Many of these adoptions occurred following placement in foster care, as opposed to infant adoption. This probably reflects the challenges parents face when taking care of children with behavioral characteristics that are difficult to manage, and it perhaps suggests that the biological parents themselves may have had cognitive, psychiatric, and/or executive-functioning challenges that made it impossible for them to provide adequate care for these children.
Finally, it is important to consider whether any of the common behavioral phenotypes observed in this study, such as intellectual disability or ASD, may be modifiable or serve as good outcome measures in future treatment studies. For instance, recent work has suggested that alpha-7 nicotinic receptor agonists may be beneficial for treating the behavioral problems prevalent in ASD25,26 and perhaps even for enhancing cognition.27 This is intriguing given the previously discussed role of the nicotinic receptor CHRNA7 in the 15q13.3 phenotype. However, it is important to consider that even if pharmacologic interventions were effective for these aspects of the phenotype, currently available assessment tools such as those used here (e.g., ADOS-2 and ADI-R) are not particularly well suited for measuring short-term changes in these phenotypes. Before therapeutic trials are attempted, it will be important to reassess a group of patients with 15q13.3 deletions using measures that are better suited to quantifying improvement from pharmacologic therapy.
We recognize several limitations of the study. First, the age range of study participants is narrow (12–18 years). Systematic neurobehavioral assessments of younger children with this disorder are warranted to better understand the developmental trajectories throughout childhood and would be meaningful in order to provide better anticipatory guidance for affected families. Second, study participants were enrolled based on a previously existing genetic diagnosis of 15q13.3 microdeletion, which represents an important ascertainment bias. Clinical CMA is considered a first-tier test in the evaluation of individuals with developmental delay, intellectual disability, congenital anomalies, or autism spectrum disorder. Therefore, individuals with respective phenotypes are inherently overrepresented in any study that enrolls on the basis of previously identified CNVs. The findings presented in this article are therefore probably not fully representative of all individuals with 15q13.3 microdeletion in the general population.
In summary, this report represents the first systematic neurobehavioral assessment of individuals with heterozygous 15q13.3 deletions and suggests that the complexity and heterogeneity of the phenotype are likely to pose a major challenge to patients, caretakers, and medical providers. Future work to more clearly define the specific contribution of the genes and noncoding RNAs in the region to various aspects of the clinical phenotype, and to understand their interactions and biological functions, will be important to advance the basic understanding of how this unique region contributes to human brain development as well as to guide development of targeted therapies.
Disclosure
The authors declare no conflict of interest.
References
Pujana MA, Nadal M, Guitart M, Armengol L, Gratacòs M, Estivill X. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur J Hum Genet 2002;10:26–35.
Chamberlain SJ, Lalande M. Neurodevelopmental disorders involving genomic imprinting at human chromosome 15q11-q13. Neurobiol Dis 2010;39:13–20.
Szafranski P, Schaaf CP, Person RE, et al. Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: benign or pathological? Hum Mutat 2010;31:840–850.
van Bon BW, Mefford HC, Menten B, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet 2009;46:511–523.
Williams NM, Franke B, Mick E, et al. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am J Psychiatry 2012;169:195–204.
Coe BP, Witherspoon K, Rosenfeld JA, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet 2014;46:1063–1071.
Lepichon JB, Bittel DC, Graf WD, Yu S. A 15q13.3 homozygous microdeletion associated with a severe neurodevelopmental disorder suggests putative functions of the TRPM1, CHRNA7, and other homozygously deleted genes. Am J Med Genet A 2010;152A:1300–1304.
Liao J, DeWard SJ, Madan-Khetarpal S, Surti U, Hu J. A small homozygous microdeletion of 15q13.3 including the CHRNA7 gene in a girl with a spectrum of severe neurodevelopmental features. Am J Med Genet A 2011;155A:2795–2800.
Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet 2008;40:322–328.
Lowther C, Costain G, Stavropoulos DJ, et al. Delineating the 15q13.3 microdeletion phenotype: a case series and comprehensive review of the literature. Genet Med 2015;17:149–157.
Ben-Shachar S, Lanpher B, German JR, et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet 2009;46:382–388.
Masurel-Paulet A, Andrieux J, Callier P, et al. Delineation of 15q13.3 microdeletions. Clin Genet 2010;78:149–161.
Miller DT, Shen Y, Weiss LA, et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet 2009;46:242–248.
Pagnamenta AT, Wing K, Sadighi Akha E, et al.; International Molecular Genetic Study of Autism Consortium. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet 2009;17:687–692.
Spielmann M, Reichelt G, Hertzberg C, et al. Homozygous deletion of chromosome 15q13.3 including CHRNA7 causes severe mental retardation, seizures, muscular hypotonia, and the loss of KLF13 and TRPM1 potentially cause macrocytosis and congenital retinal dysfunction in siblings. Eur J Med Genet 2011;54:e441–e445.
Stewart LR, Hall AL, Kang SH, Shaw CA, Beaudet AL. High frequency of known copy number abnormalities and maternal duplication 15q11-q13 in patients with combined schizophrenia and epilepsy. BMC Med Genet 2011;12:154.
Sahoo T, Theisen A, Rosenfeld JA, et al. Copy number variants of schizophrenia susceptibility loci are associated with a spectrum of speech and developmental delays and behavior problems. Genet Med 2011;13:868–880.
Gillentine MA, Schaaf CP. The human clinical phenotypes of altered CHRNA7 copy number. Biochem Pharmacol 2015;97:352–362.
Rosenfeld JA, Stephens LE, Coppinger J, et al. Deletions flanked by breakpoints 3 and 4 on 15q13 may contribute to abnormal phenotypes. Eur J Hum Genet 2011;19:547–554.
Shinawi M, Schaaf CP, Bhatt SS, et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat Genet 2009;41:1269–1271.
Hoppman-Chaney N, Wain K, Seger PR, Superneau DW, Hodge JC. Identification of single gene deletions at 15q13.3: further evidence that CHRNA7 causes the 15q13.3 microdeletion syndrome phenotype. Clin Genet 2013;83:345–351.
Jacquemont S, Coe BP, Hersch M, et al. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet 2014;94:415–425.
Ionita-Laza I, Xu B, Makarov V, et al. Scan statistic-based analysis of exome sequencing data identifies FAN1 at 15q13.3 as a susceptibility gene for schizophrenia and autism. Proc Natl Acad Sci USA 2014;111:343–348.
Zufferey F, Sherr EH, Beckmann ND, et al.; Simons VIP Consortium; 16p11.2 European Consortium. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J Med Genet 2012;49:660–668.
Cubells JF, Deoreo EH, Harvey PD, et al. Pharmaco-genetically guided treatment of recurrent rage outbursts in an adult male with 15q13.3 deletion syndrome. Am J Med Genet A 2011;155A:805–810.
Deutsch SI, Urbano MR, Neumann SA, Burket JA, Katz E. Cholinergic abnormalities in autism: is there a rationale for selective nicotinic agonist interventions? Clin Neuropharmacol 2010;33:114–120.
Wallace TL, Porter RH. Targeting the nicotinic alpha7 acetylcholine receptor to enhance cognition in disease. Biochem Pharmacol 2011;82:891–903.
Acknowledgements
This work was supported by Doris Duke Charitable Foundation grant 2011034. The project was supported in part by IDDRC grant 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). C.S. is generously supported by the Joan and Stanford Alexander Family. M.N.Z. was supported by the Medical Scientist Training Program at Baylor College of Medicine and the NICHD. We thank the patients and their families for participating in this study.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Tables
(XLS 65 kb)
Rights and permissions
About this article
Cite this article
Ziats, M., Goin-Kochel, R., Berry, L. et al. The complex behavioral phenotype of 15q13.3 microdeletion syndrome. Genet Med 18, 1111–1118 (2016). https://doi.org/10.1038/gim.2016.9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.9
Keywords
This article is cited by
-
A review of the cognitive impact of neurodevelopmental and neuropsychiatric associated copy number variants
Translational Psychiatry (2023)
-
Impaired OTUD7A-dependent Ankyrin regulation mediates neuronal dysfunction in mouse and human models of the 15q13.3 microdeletion syndrome
Molecular Psychiatry (2023)
-
Altered neuronal physiology, development, and function associated with a common chromosome 15 duplication involving CHRNA7
BMC Biology (2021)
-
Genetic copy number variants, cognition and psychosis: a meta-analysis and a family study
Molecular Psychiatry (2021)
-
First report of two successive deletions on chromosome 15q13 cytogenetic bands in a boy and girl: additional data to 15q13.3 syndrome with a report of high IQ patient
Molecular Cytogenetics (2019)