Abstract
Purpose:
While the diagnostic success of genomic sequencing expands, the complexity of this testing should not be overlooked. Numerous laboratory processes are required to support the identification, interpretation, and reporting of clinically significant variants. This study aimed to examine the workflow and reporting procedures among US laboratories to highlight shared practices and identify areas in need of standardization.
Methods:
Surveys and follow-up interviews were conducted with laboratories offering exome and/or genome sequencing to support a research program or for routine clinical services. The 73-item survey elicited multiple choice and free-text responses that were later clarified with phone interviews.
Results:
Twenty-one laboratories participated. Practices highly concordant across all groups included consent documentation, multiperson case review, and enabling patient opt-out of incidental or secondary findings analysis. Noted divergence included use of phenotypic data to inform case analysis and interpretation and reporting of case-specific quality metrics and methods. Few laboratory policies detailed procedures for data reanalysis, data sharing, or patient access to data.
Conclusion:
This study provides an overview of practices and policies of experienced exome and genome sequencing laboratories. The results enable broader consideration of which practices are becoming standard approaches, where divergence remains, and areas of development in best practice guidelines that may be helpful.
Genet Med advance online publication 03 Novemeber 2016
Similar content being viewed by others
Introduction
Exome sequencing is rapidly gaining acceptance as a useful diagnostic test in clinical medicine.1,2,3 Although a less frequently utilized option, genome sequencing enables more comprehensive genome analysis with expanded data and uniform depth of coverage, albeit at higher costs and lower average depth of coverage.4,5 With each of these tests, collectively referred to here as “genomic sequencing,” there are numerous laboratory processes required to support the identification, interpretation, and reporting of variants that may be clinically significant for the patient.
A typical genome has approximately 3.5 million differences when compared to the reference genome, of which 0.6 million are rare or novel.6 Although exome sequencing focuses on the subset of variants within or near coding sequences, hundreds to thousands of variants are identified for analysis per patient for clinical relevance.7 A major challenge is determining which, if any, of the identified variants may be relevant to the indication for testing and thereby warrant inclusion in the final test report. Beyond variants of possible diagnostic or therapeutic relevance, laboratories may also choose to identify incidental or secondary variants unrelated to the testing indication (referred to below as “secondary findings”) but potentially relevant to the patients and the health of their families. Unquestionably, the inclusion or exclusion of variants in test reports may have substantial impact on patient care. It is therefore critical to explore how these decisions are made.
Laboratories and professional societies have begun developing and recommending approaches to support this complex and labor-intensive process that combines the practice of medicine with burgeoning next-generation sequencing (NGS) laboratory procedures.8,9,10,11,12 Here, we present the results from a survey of laboratories experienced in genomic sequencing to explore protocols supporting the testing, interpretation, and reporting processes. The goal of this study was to identify workflow and reporting practices that are shared and/or discordant among laboratories in an effort to highlight practices that are becoming standard and to determine areas in need of standardization or best practice recommendations.
Materials and Methods
Recruitment
US laboratories that are Clinical Laboratory Improvement Amendments (CLIA)-certified and offer exome and/or genome sequencing—either to support the National Human Genome Research Institute (NHGRI) and National Cancer Institute (NCI)-funded Clinical Sequencing Exploratory Research (CSER) program or for routine clinical services—were invited to participate in the survey. Clinical services outside of CSER were identified primarily through a search of NCBI’s Genetic Test Registry (PMCID: PMC5005464). A total of 27 laboratory groups were identified. An invitation letter describing the purpose of the survey was sent by e-mail to each laboratory. One follow-up invitation was sent to nonresponding laboratories. Of the 27 laboratories contacted, 21 responded and completed participation (a 78% response rate) ( Table 1 ).
Participation was completely voluntary, and laboratories did not receive compensation for completion of the study. This study was submitted to the Partners HealthCare IRB for review on 12 October 2014 and was determined to be exempt. Consent was implied by agreement to participate.
Surveys
A 73-item survey was developed by the study team with input from the CSER Actionability and Return of Results Working Group (see Supplementary Materials and Methods online). Items were formatted as multiple-choice with the option to provide free text; they were grouped topically and included the following elements of genomic sequencing laboratory processes:
Consent
Sample (trio versus proband-only)
Phenotype collection
Indication-specific gene list development and use
Credentials of staff for various roles
Case review, group discussion, and clinician input
Time required for variant and case review
Sanger confirmation
Provision of gene/region coverage statistics
Secondary findings policy
Reanalysis of data
Return of raw data policies
The survey was e-mailed to each laboratory group that identified staff who were key informants regarding the laboratory’s sequencing, interpretation, and reporting processes. Surveys were completed between December 2014 and February 2015. Study leaders (H.M.M., J.M.O., and H.L.R.) reviewed the completed survey through a follow-up phone call with each laboratory’s primary respondents to clarify questions and answers. Each follow-up call lasted approximately 1 h and was audiorecorded.
Data analysis
Because of the small sample size, only frequency counts and descriptive results were utilized to demonstrate trends of agreement or divergence among the CSER program and clinical service laboratories. Counts are combined for clinical and CSER laboratories when responses do not appear to differ between the settings.
Results
Participants
Responses were collected from CLIA-certified laboratories offering exome and genome sequencing, including 9 of the 10 laboratories supporting CSER research programs as well as 12 laboratories offering routine clinical genomic services, for a total of 21 laboratories. All responses represented practices at the time of survey completion. Some laboratories supported both a routine clinical service and a CSER program. However, these laboratories usually had separate protocols (and in some cases different personnel) specific to each purpose. For the purposes of this study, these approaches were treated as two separate laboratories. Hereafter, the laboratories offering routine clinical services are referred to as “clinical” laboratories (n = 12) and the laboratories supporting research programs are referred to as “CSER” laboratories (n = 9) ( Table 1 ).
Overall, 16 laboratories (out of 21) reported a history of completing >50 exome or genome sequencing tests, and a high proportion of clinical laboratories (8 of the 12) had completed >200 tests at the time of the survey.
Sample and consent
All laboratories required written documentation of consent prior to performing the test, with some variance in whether it was signed by the patient only (12 of 21) or by the patient and physician (9 of 21).
The majority of clinical laboratories (8 of 12) performed genome-scale or exome-scale testing on trio samples in >50% of cases, whereas the opposite was true for CSER laboratories, with 6 of 9 sequencing probands only.
Phenotype collection and target gene list development
Phenotypic information was collected from the referring clinician through several means, the most frequent of which were free-text fields on the requisition form (14 of 21), attached medical records and clinic notes (14 of 21), and written responses to targeted questions (13 of 21). Only one CSER laboratory did not collect any phenotypic information, owing to their primary project aim of determining carrier status rather than diagnostic analysis.
There was notable variance in how frequently laboratories reported using indication-specific gene lists for targeted analysis of individual cases: 10 of the 21 laboratories used them in all or almost every case (>90–100%); 4 used them in the majority (50–90%) of cases; 1 used them in 10–50% of cases; and 6 rarely (<10%) or never used them. For three of the laboratories, the same gene list was applied for all cases due to the test indication: carrier status analysis or oncology (somatic and germ-line) testing. Most laboratories that used indication-based gene lists did not have a policy governing how lists were developed or updated, although two clinical laboratories reported that gene lists were assembled on a case-by-case basis.
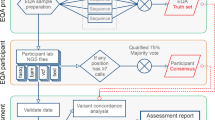
Case review
Variant interpretation and case review processes include several steps, and laboratories identified individuals with various roles in these processes ( Figure 1 ). For the purpose of this survey, we defined the initial analysis as the nonautomated primary collection of variant-level evidence for case-specific clinical interpretation. This initial analysis was most frequently performed by PhD-level analysts or fellows (medical genetics or pathology); genetic counselors were also frequently involved. Although some laboratories involved American Board of Medical Genetics and Genomics (ABMGG) board-certified medical geneticists (PhD trained) or pathologists in the initial analysis steps, these individuals were most often utilized during secondary review and report sign-out.
Group discussion was frequently used within the process of variant review for potential case-specific relevance, and all but one laboratory had regularly scheduled meetings for this purpose. This clinical laboratory instead conducted ad hoc meetings for an estimated 10–50% of cases. Approximately 60% of laboratories (12 of 21) reported that group discussion is part of all cases or almost every case (90–100%) (six clinical and six CSER). However, three clinical laboratories utilized group discussion for only a small portion (<10%) of cases. The professionals in the group discussions nearly always comprised board-certified medical and clinical geneticists, genetic counselors, and bioinformaticians, but it also commonly included basic science researchers, nongenetics physicians, trainees, and ethicists.
Exclusive of group discussions, more than one individual was involved in the interpretation and reporting of each case: seven laboratories (four clinical; three CSER) typically utilized two individuals, nine laboratories (four clinical; five CSER) utilized three individuals, and five laboratories (four clinical; one CSER) reported that four or more individuals were involved in each case. No laboratories reported using only one individual per case.
The time required for full case review was based predominantly on estimates rather than timed work and varied widely among groups (see Supplementary Materials and Methods online).
Sanger confirmation
Sanger sequencing was used for orthogonal confirmation by the majority of laboratories (17 of 21) for all reported variants or for variants considered to represent possible diagnostic findings ( Figure 2 ). Only one clinical laboratory indicated that it did not confirm any reported variants. Most laboratories had protocols specifying which types of reported variants were or were not Sanger-confirmed. For example, in one laboratory, only the germ-line variants were confirmed when paired somatic/germ-line sequencing was performed. Another laboratory chose not to confirm reported variants related to pharmacogenetics, carrier status, or low-penetrant genetic risk associations. Yet another confirmed only variants that clearly matched the clinical indication question; if a particular variant(s) explained only one of several clinical features, then it would not be confirmed. Notably, the survey did not elicit analytical quality thresholds used in the decisions of which variants to review; instead, it focused on policies regarding how different types of variants are handled.
Sequencing test report
Variant classes included in the report were influenced by whether the variant was relevant to manifesting symptoms or considered a secondary finding. Regarding diagnostic indications, 19 of 20 laboratories reported variants deemed pathogenic or likely pathogenic as well as variants of uncertain significance (VUS) if in a gene related to manifesting symptoms. None of the clinical laboratories returned VUSs for secondary findings; however, two CSER laboratories did, which is consistent with their research goals. Three laboratories included likely benign or benign variants on the test report for diagnostic findings.
Case-specific filtering strategies, including gene lists, were used by most laboratories during analysis for at least a portion of cases to facilitate identification of potentially relevant genes/variants. In addition, case-specific data quality and exon coverage for phenotypically relevant genes may be considered during case analyses. Interestingly, 4 of 15 laboratories that reported the use of phenotypically derived gene lists in at least a portion of cases indicated that such methods were not regularly communicated in the test report, although one laboratory would make them available by request. Further, case-specific coverage of genes, regardless of whether a gene list was used, was not universally conveyed. Ten of 21 laboratories routinely reported case-specific coverage, and an additional 4 laboratories made it available separately or by request.
Secondary findings
All laboratories indicated that they report secondary findings; however, there was considerable variability in the types of secondary findings reported and whether patients were allowed to “opt out” of receiving them ( Table 2 ). Eight clinical laboratories allowed patients to opt out of receiving these results, whereas four of the clinical laboratories required “opt-in” for secondary findings, reflecting deliberate patient choice. Of the CSER laboratories, one targeted to healthy participants did not allow opt-out of any findings, seven allowed opt-out of all secondary findings, and one required disclosure for a predefined set of genes while requiring opt-in to learn additional subsets. It is noted that the specific aims of each study influenced CSER laboratory approaches toward return of secondary findings and that the CSER protocols were likely written prior to the publication of the ACMG guidelines.12,13
Five clinical laboratories limited secondary finding variant analysis and reporting principally to the 56 genes recommended for return by the American College of Medical Genetics and Genomics (ACMG),12 although three of these reported additional “medically actionable findings” incidentally discovered in the course of the primary diagnostic analysis. Another clinical laboratory reported variants within the ACMG 56 genes only if they were discovered in the course of the primary diagnostic analysis. The number of genes included for analysis and return ranged from the ACMG 56 to “any human disease gene” (>4,500 genes) and included categories such as monogenic disease, carrier status, pharmacogenomic variants, complex traits, and blood antigen prediction. Again, research aims influenced the number and categories for CSER laboratories, as exemplified by one that did not analyze four of the ACMG genes that caused childhood-onset conditions (RB1, WT1, APOB, and PCSK9) because their study enrolled only adults but added 50 additional adult-onset medically actionable genes to their list.
Reanalysis
The majority of laboratories indicated that they had reanalyzed case-specific data to provide an updated report at least once (11 of 12 clinical and 4 of 9 CSER). The instances were rare, however, with 7 of 12 clinical and 6 of 9 CSER laboratories indicating that reanalysis rarely or never occurred. Only one clinical laboratory routinely reanalyzed every case. When reanalysis was performed, approximately half used the existing variant call format (VCF) file and half performed new alignment and variant calling. Of the clinical laboratories, six indicated that reanalysis would be free of charge, five charged a fee, and one was still developing its policy.
Raw data return
The US Department of Health and Human Services issued a rule change that went into effect in April 2014, specifying that clinical laboratories must provide copies of completed test reports to the patient upon request.14 At the time of survey collection, there was some question regarding what types of data this encompassed.15 When queried about this new rule, three clinical laboratories indicated they would return uninterpreted sequencing data to patients and/or physicians upon request, whereas eight clinical laboratories would return it only to the physician. However, the frequency of actually doing this was low; one clinical laboratory had returned such data to patients (<10% of cases). Return to physicians was more frequent, with four returning it in 10–50% of cases and four returning it for fewer than 10% of cases. Laboratories indicated that a fee may be charged to offset costs associated with storage devices, shipping, and other expenses.
Discussion
The results of our exploratory survey revealed numerous areas of convergence across laboratories, suggesting practices that may represent a developing standard for exome and genome sequencing. Examples include consent documentation, default inclusion of some secondary findings as well as allowing some choice, and case review by more than one individual. There were also some notable areas of divergence that warrant further discussion and possible clarification through practice guideline development.
Clinical NGS guidelines recommend detailed phenotypic information to aid the laboratory analysis and interpretation process.10 Consistent with this recommendation, phenotypic data about the clinical indication(s) for testing were collected by all laboratories in various formats. The creation of phenotype-guided gene lists for defining high-priority genes was not uniformly used during the bioinformatic filtering steps. Although many cases may be solved without such gene lists, the prioritized review of variants in genes most likely to be implicated enables further in-depth consideration of variants in higher-likelihood genes to ensure that they are not overlooked. The absence of these approaches in some laboratories may reflect the diversity of bioinformatics pipelines used or other strategies to incorporate phenotype. It is also worth noting that if phenotypic assumptions are incorrect, then overreliance on gene lists could cause a clinically relevant variant to be missed. Continued standardization of analysis pipelines, robust methods and standardized ontologies for phenotypic collection, and the development of high-quality curated gene–phenotype data sets are important to ensure comprehensive, consistent case analysis.
In conflict with sequencing guidelines, case-specific analysis approaches (e.g., phenotypically guided gene lists) were not routinely communicated through the test report.10 When these analysis details are omitted from the report, it may be unclear to the clinician which genes were or were not analyzed for their patient or how well any particular gene was covered. This information would be most important in the case of a negative result. Failing to provide a clear depiction of gene-centric analysis and coverage could lead to false assumptions by the ordering clinician that key genes had been ruled out.
Another area of discordance was revealed in laboratories’ use of group discussion in the case review process. Although all laboratories reported that more than one individual was involved in the analysis and interpretation of a given case, only slightly more than half routinely utilized group review for most cases. Benefits of group discussion or solicitation of ad hoc expertise include the insight from multiple perspectives and expertise, including bioinformatics,8 basic science, and clinical domain knowledge.16 This “peer review” may improve accuracy and confidence in decision making regarding the potential clinical relevance of variants.17 Case volume, time demands, and costs are obvious constraining factors for implementing group case review, as is solicitation of disease-area expertise. It may be practical for experienced laboratories to consider this approach only for particularly challenging cases.
Most laboratories were in agreement about which variant classification categories warranted inclusion on the clinical report. However, policies governing the confirmation of reported variants, typically through Sanger sequencing, differed somewhat among laboratories. Notably, one clinical laboratory indicated that it does not confirm any variants, and several others had certain categories of reported variants that were not confirmed. Although it has generally been recommended that laboratories Sanger-confirm reported germ-line variants,10 as laboratories gain experience with the analytic performance of NGS, there is increasing movement toward defining thresholds for quality (and perhaps clinical significance) for which such confirmation is unnecessary.18,19,20
Survey results indicated that reanalysis of data remains predominantly an ad hoc service performed upon request rather than an integrated process. Several factors may play a role in this absence of common practice, including the rapid pace at which new genetic knowledge is generated as well as the relatively new addition of exome and genome testing services (many clinical exome sequencing services are less than 3 years old). Another important driver is lack of the billing reimbursement infrastructure needed to support data reanalysis, interpretation, and reporting.21
In 2014, new federal rules issued by the US Department of Health and Human Services (DHHS) specified that clinical laboratories are required to provide copies of completed test reports directly to the patient upon request.14 January 2016 guidance from DHHS clarified that for genomic tests, the access right includes “a copy of the completed test report, the full gene variant information generated by the test, as well as any other information in the designated record set concerning the test.”22 A major aim of the rule and guidance is to grant individuals access to their protected health information maintained by providers. Probably due in part to uncertainty regarding the rule, the majority of laboratories reported that they would release uninterpreted or unvalidated sequence data to the ordering physician only. Despite the recent 2016 clarification, guidance regarding how best to effectuate this right of access may be welcomed by the laboratory community. The customary practice in medicine has been to convey medical data through a patient’s clinician, who can interpret its significance in the appropriate clinical context, because uninterpreted and unvalidated data would probably be inaccessible to direct interpretation by most patients. Laboratory practices are likely to continue to evolve as patients are taking an increasingly active role in their health and exercising their right to advance medicine through data sharing. Laboratories and clinicians may perceive some risks in data return that are both profession-related (if subsequent analyses were to conflict with the original assessment) and resource-related. It is, however, critical that we, as professionals, maintain transparency and support peer review of our practices to ensure that the highest quality care is delivered to patients.
Limitations
Given the rapid evolution of genomic sequencing practices, the testing market, and legal rules, as well as the <100% response from all laboratories at the time of this survey, these results may not reflect current exome and genome sequencing practices. However, all respondents reviewed the data for accuracy and major changes immediately prior to submission. In addition, these results do not reflect practices outside the United States, and the majority of laboratories represented were primarily academic institutions, many of which may have had practices influenced by specific grant funding for clinical sequencing. Furthermore, because the survey review was conducted by respondents’ professional peers, it is possible that some responses may have been impacted by perceived best practices. Finally, this study provides observational data about current practices. There remains a need, however, for systemic evidence collection to define the clinical validity and utility of genomic sequencing such that recommended best practices can be better informed by underlying evidence.
Conclusions
This study provides an overview of general practices and policies of experienced exome and genome sequencing laboratories. The results enable broader consideration of which practices are becoming standard approaches and where development of best practice guidelines may be helpful. Notable areas for improvement include the following:
-
1
(i) Transparency and clarity regarding test methods and limitations. We recommend that the scope of analysis (including the use of gene lists), case-specific coverage metrics, and analytical limitations, be communicated with the report. Information might include:
-
1
(i) A list of genes targeted for analysis and the phenotype elements used to select them
-
2
(ii) The threshold for minimum coverage and notation when coverage of a targeted gene falls below that threshold and/or
-
3
(iii) Known pathogenic variation relevant to the indication but not detectable by the test
-
1
-
2
2. Utilization of clinical domain expertise in case review. We suggest that laboratories consider implementing group case review with inclusion of varied expertise including clinical domain expertise. Because group review is not necessary for all cases, laboratories may wish to define circumstances in which such a review will improve case-specific determination of clinically relevant variants to report as well as provide a rich learning environment for all staff.
-
3
3. Confirmation of reported variants. We recommend that all reports of variants with potential diagnostic significance meet a defined threshold for data quality or require confirmation by an orthogonal method. This should apply to both indication-specific and secondary finding variants likely to be used in clinical care. This would not apply to the return of uninterpreted sequence data addressed below under “Data access guidelines.”
-
4
4. Data access guidelines. Federal rules and guidance now establish a patient’s right of access to “the completed test report, the full gene variant information generated by the test, as well as any other information in the designated record set concerning the test.” We suggest that the professional community establish guidelines surrounding the return of sequencing data directly to patients to guide laboratories in honoring patients’ right of access while minimizing potential harm from misunderstood or incorrectly interpreted results.
-
5
5. Data reanalysis. We recommend that laboratories develop internal genomic sequencing data reanalysis guidelines. These guidelines are best informed by the professional community and should address both laboratory-initiated reanalysis and clinician-initiated reanalysis as well as the appropriate data to be reanalyzed (existing VCF vs. raw data requiring new alignment and base calling vs. new sequencing run) and when retesting would be recommended over reanalysis.
Disclosure
Most authors are clinical service providers and many are employed by laboratories which offer fee-based clinical sequencing. This employment is noted in the author affiliations. Additional commercial conflict of interest beyond any author's employment affiliation include: A.S. (Invitae (Scientific Consultant), Agilent (licensing)) S.J.B. (GeneDx, BioReference (stock), advisory boards for RainDance, Ingenuity). The other authors declare no additional conflict of interest.
References
Retterer K, Juusola J, Cho MT, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med 2016;18:696–704.
Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014;312:1870–1879.
Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 2014;312:1880–1887.
Belkadi A, Bolze A, Itan Y, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA 2015;112:5473–5478.
Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics 2014;15:247.
Kohane IS, Hsing M, Kong SW. Taxonomizing, sizing, and overcoming the incidentalome. Genet Med 2012;14:399–404.
McLaughlin HM, Ceyhan-Birsoy O, Christensen KD, et al.; MedSeq Project. A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet 2014;15:134.
Gargis AS, Kalman L, Bick DP, et al. Good laboratory practice for clinical next-generation sequencing informatics pipelines. Nat Biotechnol 2015;33:689–693.
Richards S, Aziz N, Bale S, et al.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424.
Rehm HL, Bale SJ, Bayrak-Toydemir P, et al.; Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Commitee. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013;15:733–747.
MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature 2014;508:469–476.
Green RC, Berg JS, Grody WW, et al.; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–574.
Berg JS, Amendola LM, Eng C, et al.; Members of the CSER Actionability and Return of Results Working Group. Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research Consortium. Genet Med 2013;15:860–867.
US Dept of Health and Human Services, Center for Medicare & Medicaid. CLIA program and HIPAA privacy rule: patients’ access to test reports.Final rule. Fed Regist 2014;79:7289–316.
Evans BJ, Dorschner MO, Burke W, Jarvik GP. Regulatory changes raise troubling questions for genomic testing. Genet Med 2014;16:799–803.
Brownstein CA, Beggs AH, Homer N, et al. An international effort towards developing standards for best practices in analysis, interpretation and reporting of clinical genome sequencing results in the CLARITY Challenge. Genome Biol 2014;15:R53.
Amendola LM, Dorschner MO, Robertson PD, et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res 2015;25:305–315.
Strom SP, Lee H, Das K, et al. Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory. Genet Med 2014;16:510–515.
Baudhuin LM, Lagerstedt SA, Klee EW, Fadra N, Oglesbee D, Ferber MJ. Confirming variants in next-generation sequencing panel testing by Sanger sequencing. J Mol Diagn 2015;17:456–461.
Beck TF, Mullikin JC, Biesecker LG ; NISC Comparative Sequencing Program. Systematic evaluation of Sanger validation of next-generation sequencing variants. Clin Chem 2016;62:647–654.
Deverka PA, Dreyfus JC. Clinical integration of next generation sequencing: coverage and reimbursement challenges. J Law Med Ethics 2014;42 Suppl 1:22–41.
US Dept of Health and Human Services. Individuals’ Right under HIPAA to Access their Health Information 45 CFR § 164.524. http://www.hhs.gov/hipaa/for-professionals/privacy/guidance/access/index.html. Accessed March 2016.
Acknowledgements
This work was conducted by the authors as a project guided by the Actionability and Return of Results work group of the Clinical Sequencing Exploratory Research (CSER) program. We thank all members of the work group and of the laboratories for their contributions to this project and article. We are also grateful to Joseph Salama and Ragan Hart for their assistance coordinating phone interviews. A full list of CSER consortium work group members can be found in the Supplementary Materials and Methods online. Research reported in this publication was supported by the National Human Genome Research Institute, in conjunction with additional funding from the National Cancer Institute, under awards U01HG006500, U01HG006507, U01HG007307, U41HG006834, R01HG006600, U01HG006487, UM1HG007301, U01HG006546, U01HG006492, UM1HG006508, U01HG006485 R01HG006600, UM1HG007292, and R01CA154517. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Materials
(DOCX 246 kb)
Rights and permissions
About this article
Cite this article
O’Daniel, J., McLaughlin, H., Amendola, L. et al. A survey of current practices for genomic sequencing test interpretation and reporting processes in US laboratories. Genet Med 19, 575–582 (2017). https://doi.org/10.1038/gim.2016.152
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2016.152