Abstract
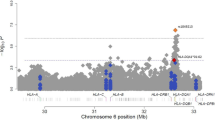
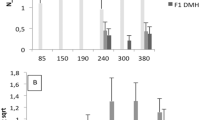
We tested the possibility to map loci affecting the acute inflammatory response (AIR) in an (AIRmax × AIRmin) F2 intercross mouse population derived from non-inbred parents, by association analysis in the absence of pedigree information. Using 1064 autosomal single nucleotide polymorphisms (SNPs), we clustered the intercross population into 12 groups of genetically related individuals. Association analysis adjusted for genetic clusters allowed to identify two loci, inflammatory response modulator 1 (Irm1) on chromosome 7 previously detected by genetic linkage analysis in the F2 mice, and a new locus on chromosome 5 (Irm2), linked to the number of infiltrating cells in subcutaneous inflammatory exudates (Irm1: P=6.3 × 10−7; Irm2: P=8.2 × 10−5) and interleukin 1 beta (IL-1β) production (Irm1: P=1.9 × 10−16; Irm2: P=1.1 × 10−6). Use of a polygenic model based on additive effects of the rare alleles of 15 or 18 SNPs associated at suggestive genome-wide statistical threshold (P<3.4 × 10−3) with the number of infiltrating cells or IL-1β production, respectively, allowed prediction of the inflammatory response of progenitor AIR mice. Our findings suggest the usefulness of association analysis in combination with genetic clustering to map loci affecting complex phenotypes in non-inbred animal species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ibanez OM, Stiffel C, Ribeiro OG, Cabrera WK, Massa S, De Franco M et al. Genetics of nonspecific immunity: I. Bidirectional selective breeding of lines of mice endowed with maximal or minimal inflammatory responsiveness. Eur J Immunol 1992; 22: 2555–2563.
Biozzi G, Ribeiro OG, Saran A, Araujo ML, Maria DA, De Franco M et al. Effect of genetic modification of acute inflammatory responsiveness on tumorigenesis in the mouse. Carcinogenesis 1998; 19: 337–346.
Vorraro F, Galvan A, Cabrera WH, Carneiro PS, Ribeiro OG, De Franco M et al. Genetic control of IL-1beta production and inflammatory response by the mouse Irm1 locus. J Immunol 2010; 185: 1616–1621.
Gao X, Starmer JD . AWclust: point-and-click software for non-parametric population structure analysis. BMC Bioinformatics 2008; 9: 77.
Darvasi A, Weinreb A, Minke V, Weller JI, Soller M . Detecting marker-QTL linkage and estimating QTL gene effect and map location using a saturated genetic map. Genetics 1993; 134: 943–951.
Talbot CJ, Nicod A, Cherny SS, Fulker DW, Collins AC, Flint J . High-resolution mapping of quantitative trait loci in outbred mice. Nature Genet 1999; 21: 305–308.
Galvan A, Vorraro F, Cabrera WH, Ribeiro OG, Pazzaglia S, Mancuso M et al. Genetic heterogeneity of inflammatory response and skin tumorigenesis in phenotypically selected mouse lines. Cancer Lett 2010; 295: 54–58.
Galvan A, Ioannidis JP, Dragani TA . Beyond genome-wide association studies: genetic heterogeneity and individual predisposition to cancer. Trends Genet 2010; 26: 132–141.
Benyamin B, Visscher PM, McRae AF . Family-based genome-wide association studies. Pharmacogenomics 2009; 10: 181–190.
Labasi JM, Petrushova N, Donovan C, McCurdy S, Lira P, Payette MM et al. Absence of the P2X7 receptor alters leukocyte function and attenuates an inflammatory response. J Immunol 2002; 168: 6436–6445.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Lander E, Kruglyak L . Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genet 1995; 11: 241–247.
Acknowledgements
This work was supported in part by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grant 03/05486-7 to OCMI and by Associazione Italiana and Fondazione Italiana Ricerca Cancro (AIRC and FIRC) grants to TAD. FV was supported in part by FAPESP, OCMI and MDF by Conselho Nacional de Pesquisa (CNPq). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Galvan, A., Vorraro, F., Cabrera, W. et al. Association study by genetic clustering detects multiple inflammatory response loci in non-inbred mice. Genes Immun 12, 390–394 (2011). https://doi.org/10.1038/gene.2011.10
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2011.10