Abstract
Spondylocostal dysostosis (SCD) is an inherited disorder with abnormal vertebral segmentation that results in extensive hemivertebrae, truncal shortening and abnormally aligned ribs. It arises during embryonic development by a disruption of formation of somites (the precursor tissue of the vertebrae, ribs and associated tendons and muscles). Four genes causing a subset of autosomal recessive forms of this disease have been identified: DLL3 (SCDO1: MIM 277300), MESP2 (SCDO2: MIM 608681), LFNG (SCDO3: MIM609813) and HES7 (SCDO4). These genes are all essential components of the Notch signalling pathway, which has multiple roles in development and disease. Previously, only a single SCD-causative missense mutation was described in HES7. In this study, we have identified two new missense mutations in the HES7 gene in a single family, with only individuals carrying both mutant alleles being affected by SCD. In vitro functional analysis revealed that one of the mutant HES7 proteins was unable to repress gene expression by DNA binding or protein heterodimerization.
Similar content being viewed by others
INTRODUCTION
The vertebral column is composed of a regular array of vertebrae, which are derived from somites in the developing embryo. The formation of somites is disrupted in abnormal vertebral segmentation, which is a common congenital abnormality.1 Abnormal vertebral segmentation can appear as uneven or fused vertebrae, present either singly or in multiples throughout the vertebral column. The term spondylocostal dysostosis (SCD) is used to describe a wide variety of radiological features that include multiple abnormal vertebral segmentation, usually with contiguous involvement of more than 10 segments, with malalignment, fusions and absence of some ribs. In recent years, genetic mapping and candidate gene sequencing approaches have identified causative mutations in four genes, all components of the Notch signalling pathway. SCDO1 (OMIM 2773002) represents the majority of cases in which the underlying genetic cause has been identified and is due to mutation of the DLL3 gene, with 27 distinct causative mutations identified to date (1, 2, 3 and unpublished). Two rarer forms are due to mutation of MESP2 (SCDO2: OMIM 6086814) and LFNG (SCDO3: OMIM 6098135); in each case, only a single SCD-causative missense mutation has been described. Finally, we recently used autozygosity mapping of a consanguineous family to identify an SCD-causative mutation in the HES7 gene.6 The functions of all these proteins are conserved during evolution, and targeted inactivation of all four genes in mouse produces phenotypes similar to each other and to SCD.7, 8, 9, 10, 11
The formation of somites from the unsegmented precursor tissue, presomitic mesoderm, is tightly controlled at the molecular level by interaction of several signal-transduction pathways including FGF, Wnt and Notch (reviewed in Ref. 12). In this process, the Notch signalling pathway is activated in the presomitic mesoderm in regular pulses, with a period equal to the time required for the formation of a single somite. This leads to the periodic activation of Notch target genes such as LFNG and HES7 in the presomitic mesoderm. HES7 is a member of the Hairy-and-enhancer-of-split (HES) family of transcription factors.13, 14 These proteins belong to the basic-helix-loop-helix (bHLH) superfamily of more than 125 DNA-binding transcription factors that regulate many key biological processes in vertebrates, invertebrates and plants. In HES proteins, the basic portion of the domain is required for DNA binding to N-box sequences (5′-CACNAG-3′), and the HLH portion is required for homo- and heterodimerization between family members. They also have a conserved domain immediately carboxy-terminal to the bHLH domain (the Orange domain), and a carboxy-terminal tetrapeptide (WRPW in HES and YRPW in the related HEY family of proteins) that mediates repression of HES target genes. In vivo, HES7 inhibits its own expression, as well as that of glycosyltransferase Lfng, and thereby indirectly inhibits Notch signalling.15 In somitogenesis, HES7 is expressed in an oscillatory manner, and mouse knockout experiments reveal that it is absolutely required for correct somite formation.7
MATERIALS AND METHODS
Human Subjects
Appropriate informed consent was obtained from the proband and family for this study.
DNA Sequencing
Genomic DNA was amplified by PCR using Herculase proofreading polymerase (Stratagene, Santa Clara, USA) and was gel purified using QIAquick purification columns (Qiagen, Hilden, Germany). Both strands were sequenced using a BigDye terminator Cycle Sequencing kit (PE Biosystems, Carlsbad, CA, USA) according to the manufacturer's instructions. Reactions were analyzed on an ABI Prism 377 DNA Sequencer (PE Biosystems) at the UNSW DNA sequencing analysis facility. HES7-specific primers were as previously described.6
Transcription Assays
Mouse muscle satellite C2C12 cells were grown in DMEM (Gibco-BRL, Carlsbad, CA, USA) containing 10% FCS (Sigma, St Louis, MO, USA). Transfections were performed using LipofectAMINE/PLUS reagent (Invitrogen, Carlsbad, CA, USA) in 12-well trays. Cells were co-transfected with plasmids encoding wild-type or mutant Hes7, E47 (for the E-box assay), and firefly (6 × N-box beta-actin promoter in Figure 3a, c or 7 × E-box beta-actin promoter in Figure 3b) and Renilla (SV40 promoter) luciferase reporters. Cells were harvested in 250 μl of passive lysis buffer (Promega, Madison, WI, USA) 24 h after transfection. Firefly and Renilla luciferase activities were assayed using the Dual-Luciferase reporter system (Promega) and measured on a FLUOstar Optima Luminometer (BMG Labtech, Offenburg, Germany). Firefly luciferase counts were normalised against Renilla luciferase counts to account for differences in transfection efficiency. One-way analysis of variance was performed on data from four independent experiments. Significance was determined using Tukey's post hoc test.
Functional analysis of I58V and D186Y mutations. (a) Wild-type and I58V Hes7 significantly repress transcription from a beta-actin promoter with six copies of an N-box sequence upstream, whereas D186Y Hes7 has significantly reduced activity. ***P<0.0001; ns, not significant. (b) Wild-type and I58V Hes7 significantly repress transcription from a beta-actin promoter with seven copies of an E-box sequence upstream, whereas D186Y Hes7 has significantly reduced activity. ***P<0.0001; ns, not significant. (c) Heterodimerization of wild-type or I58V Hes7 in combination with D186Y Hes7 rescues wild-type levels of transcriptional repression of a beta-actin promoter with six copies of an N-box sequence upstream. The reduced repression level of D186Y Hes7 expressed alone was significantly different from each of the other combinations. **P<0.01. Mouse muscle satellite C2C12 cells were co-transfected with plasmids encoding wild-type or mutant Hes7, E47 (in b), and firefly (6x N-box beta-actin promoter in a, c or 7x E-box beta-actin promoter in c and Renilla (SV40 promoter) luciferase reporters. Assays were performed in triplicate. Error bars represent standard deviations of four independent experiments. One-way analysis of variance was performed on data from all four experiments, and significance was determined using Tukey's post hoc test.
RESULTS
We investigated a family of southern European origin with two offspring affected with SCD. Consanguinity was deemed unlikely, as the parents originated from geographically separated populations in Northern and Southern Italy. The first affected individual (II.1) was born at 41 weeks gestation, with a birth weight of 3400 g (75th centile) and length of 44 cm (<3rd centile). Newborn radiographs showed multiple segmentation anomalies and rib fusions (Figure 1a). She had a short trunk and short stature, and during the first year, it was noted that her span was greater than her length. Apart from a short neck and loss of thoracic (hypokyphosis) and lumbar (hypolordotic) curves, she developed normally with normal intelligence. Clinical examination showed a short broad neck with no lateral movement, but with a small amount of rotation and flexion in the anterior/posterior plane. There was limited trunk movement throughout the spine, although there was a small degree of rotation and forward flexion of the trunk in the lumbar spine. During childhood, she developed scoliosis convex to the left, centred about T10, with a compensatory curve in the lumbar spine centred on L3. She has 11 pairs of ribs on both sides with lateral fusion of the right first and second and third and fourth ribs. On the left, the fifth and sixth ribs are fused posteriorly. An MRI at 21 years of age (Figure 1b) showed multiple segmentation anomalies in the cervical spine with butterfly and hemivertebrae. There is a sagittal cleft throughout the body of C2, and a defect (spina bifida) of the anterior ring of C1, with absence of ossification of the posterior ring. The arch of C1 is fused to C2 anteriorly. Some spinous processes are fused. There is superior displacement of the odontoid process. The junction of the medulla and cervical spinal cord is angulated anteriorly. Magnetic resonance angiography showed a dominant right vertebral artery with hypoplasia of the left vertebral artery, which does not communicate with the basilar artery. At 23 years of age, her pulmonary function showed a forced vital capacity that was 53% of predicted and a forced expiratory volume of one second (FEV1), which was 52% of predicted. Functional residual capacity was decreased in proportion to total lung capacity, consistent with a moderate restrictive ventilatory defect. Following correction for alveolar volume, the diffusing capacity for carbon monoxide (DLco) was 88% of predicted, in the lower half of its reference range consistent with normal gas transfer.
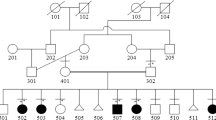
Radiograph (a) and T2-weighted coronal MRI images in the vertebral plane of individual II.1 (b) and coronal MRI image of individual II.3 (c), showing severe vertebral segmentation anomalies throughout the vertebral column. (d) Coronal view of a magnetic resonance angiography image showing the vessels of the anterior carotid and vertebrobasilar system of individual II.3 showing hypoplasia of the left vertebral artery. There is a normal right vertebral artery, and the basilar artery seems to be of normal calibre. Ica, internal carotid artery; rva, right vertebral artery; lva, left vertebral artery; ba, basilar artery.
The second affected individual (II.3) was born by normal delivery at 39 weeks gestation with a birth weight of 3160 g (50th centile) and length of 43 cm (<3rd centile). Newborn radiographs showed eight ribs on both sides, with costal fusions on the right and multiple hemivertebrae with apparent block fusion L2–L3. The upper segment to lower segment ratio was 1.55. Postnatal growth remained less than the third centile, with span greater than height. At 13 years 3 months of age, height was 140.7 cm (<3rd centile), span was 162 cm (75th centile) and the upper segment/lower segment ratio was 0.85. He developed a prominent pectus excavatum with stiffness in the trunk and neck, thoracic hypokyphosis and lumbar hypolordosis. An X-ray of the wrist and ankle at the age of 10 years showed no evidence of carpal or tarsal fusions. MRI at 15 years of age in the bone mode showed multiple segmentation anomalies in the cervical and thoracic region (Figure 1c). Magnetic resonance angiography demonstrated a hypoplastic left vertebral artery with absence of both posterior communicating arteries (Figure 1d).
Genomic DNA from individual II.3 (Figure 2a) was sequenced for the entire coding region and splice sites of the four genes previously shown to cause SCD (DLL3, MESP2, LFNG and HES73, 4, 5, 6). DLL3, MESP2 and LFNG genes showed no sequence deviations from the reference sequence. However, two heterozygous missense mutations (c.172A>G in exon 3 and c.556G>T in exon 4) were detected in HES7, resulting in substitution of valine for isoleucine (I58V) and tyrosine for aspartic acid (D186Y). Further sequencing analysis of the entire family revealed that the other affected individual (II.1) was also compound heterozygous for I58V and D186Y, whereas each parent carried only a single missense mutation (the father carrying the I58V allele and the mother carrying the D186Y allele; Figure 2a). One unaffected sibling also carried the D186Y allele, and the other was wild type at both sequence positions. Neither base change created a restriction fragment length polymorphism; hence, to confirm the presence of a bona fide sequence alteration, products from at least two independent PCR reactions were sequenced for each affected individual. To demonstrate that these base changes were not common polymorphisms unassociated with the SCD phenotype, 110 ethnically matched control subjects (220 chromosomes) were sequenced. No control chromosome contained a mutation at either of these positions, giving approximately 80% power to distinguish a normal sequence variant from a mutation.16 In addition, the underlying base substitutions were not present in the NCBI SNP database.
Detection of HES7 mutations c.172A>G and c.556G>T. (a) Pedigree of family affected with SCD, with affected status indicated by black shading. The genotypes of each individual are shown using the predicted amino-acid change. (b) Electropherograms showing the presence of c.172A>G and c.556G>T mutations in the probands, their parents and unaffected siblings. (c) Comparison of the amino-acid sequence of the human HES7 bHLH region with that of the most closely related proteins of mouse, zebrafish and Drosophila melanogaster. The arrow indicates the amino acid mutated in I58V; residues LeuH1.7, LeuH1.10 and LysH2.1 are indicated by black circles; and the conserved proline residue by an asterisk. Amino acids are numbered according to human HES7.
To investigate the effects of these base changes on HES7 function, we used two previously described cell culture transcription repression assays.17 HES family proteins repress transcription through two distinct mechanisms. These proteins bind directly to DNA through an N-box (CACNAG), using a basic region immediately amino-terminal to the helix-loop-helix domain (which is involved in homo- and heterodimerization with other bHLH family members). Co-repressors are then recruited to the promoter through interaction with a WRPW carboxy-terminal motif. They can also form heterodimers with the bHLH protein E47, thus preventing it (and other bHLH proteins that normally heterodimerize with E47 such as MyoD) from binding to E-boxes (CANNTG) and activating transcription. As no full-length human HES7 cDNA was available, we created I58W and D186Y mutations in mouse Hes7, given that their bHLH regions have the same amino-acid sequence (Figure 2b) and the entire proteins are 92% identical. Expression of wild-type Hes7 represses transcription from a beta-actin reporter with upstream N-boxes (Figure 3a), and represses E47-dependent transcription from a beta-actin reporter with upstream E-boxes (Figure 3b) in mouse muscle satellite C2C12 cells. When the D186Y mutant form of Hes7 was used in the N-box assay, transcription was repressed to a significantly lesser extent (P<0.0001) than with wild-type Hes7. In contrast, the I58V mutation showed no significant difference in the degree of repression compared with wild-type Hes7. The observed reduction in the ability of the D186Y mutant to repress transcription in the N-box assay might have been due to a reduction in protein expression level, or a destabilization of the Hes7 protein. We therefore tested this by transiently transfecting C2C12 with HA-tagged versions of wild-type and mutant Hes7 proteins. To monitor the relative protein stabilities, cells were treated with cycloheximide, and protein extracts were obtained at 0, 20, 40 and 60 min intervals. Hes7 protein levels were determined by means of western blots with beta-tubulin used as a loading control (Supplementary Figure). These experiments showed that all three proteins were expressed at similar levels under these conditions, and neither of the two mutant proteins was cleared from the cell in a manner different from that of the wild-type protein. This suggests that the D186Y mutant has reduced activity in the N-box assay due to an intrinsic disruption of its repressive activity. Similarly, in the E-box assay, the D186Y mutant Hes7 repressed to a significantly lesser extent (P<0.0001) than did wild-type Hes7, whereas the I58V mutant repressed transcription to the same degree as did wild-type Hes7. This suggests that the D186Y mutation in the carboxy-terminal region of Hes7 also impairs the ability of Hes7 to heterodimerize with E47. The I58V mutant protein is not functionally compromised in either of the repression assays. However, we noted that the SCD phenotype is only observed when both I58V and D186Y mutations are present in an individual, and this may reflect a reduced capacity of the I58V mutant protein to dimerize with D186Y Hes7. We therefore co-expressed both I58V and D186Y Hes7 mutants in the N-box repression assay, and compared the level of repression with that of cells co-expressing wild-type and D186Y Hes7; wild-type and I58V Hes7; or each Hes7 protein singly (Figure 3c). In this assay, I58V, in combination with either wild-type or D186Y Hes7, had a statistically insignificantly different repressive activity compared with wild-type Hes7, suggesting that the I58V Hes7 protein is able to dimerize with D186Y Hes7 equally efficiently as with wild-type Hes7. This experiment also shows that, when co-expressed with D186Y Hes7, an equal amount of either wild-type or I58V Hes7 is sufficient to rescue normal repressive activity in the N-box assay.
DISCUSSION
Of the four Notch pathway-associated genes implicated in causing SCD, HES7 is perhaps the most interesting because of its ability to indirectly inhibit Notch signalling in a periodic manner, driving the cyclical activation of Notch that is required for normal somite formation. In this study, we report two novel HES7 mutations (D186Y and I58V) in a nonconsanguineous family with SCD. When expressed on its own, the D186Y mutation clearly disrupts normal HES7 activity in two functional assays in vitro. This is not due to changes in the protein expression level or protein stability, and is therefore likely to be due to a disruption of the repression and/or heterodimerization properties of the protein. The D186 residue is completely conserved in all published mammalian sequences; however, no structural data are available for the proline-rich domain extending from residue 128 to the carboxy terminus. More distant vertebrates (zebrafish) and invertebrates (Drosophila) do not have an orthologue of HES7. The most closely related proteins are her1 (zebrafish) and hairy (Drosophila), which share a similar domain organization to HES7, but are very divergent outside the basic-helix-loop-helix domain. The nonconservative change from aspartic acid to tyrosine results in the replacement of a residue with a strong negative charge with a bulkier residue with only a potential partial charge; therefore, it is possible that the charge of the aspartic acid residue is required for the protein–protein interactions needed for the full repressive activity or heterodimerization potential of HES7. By contrast, the I58V mutant Hes7 showed activity that was indistinguishable from that of wild-type protein in both assays when expressed on its own, as well as when expressed in combination with wild-type or D186Y Hes7 in the N-box assay. This is perhaps surprising, as the I58V mutation alters an evolutionarily conserved hydrophobic residue in the second helix of the bHLH domain (Figure 2b). This part of the bHLH domain is required for homo- and heterodimerization of the HES7 protein. Although the precise three-dimensional structure of this part of the HES7 protein is not known, X-ray crystallographic studies of other bHLH domains can be used to extrapolate the effects of such an amino-acid substitution. Longo et al18 compared the crystal structures of the E47 homodimer and the E47-NeuroD heterodimer. In these structures, the residue homologous to I58 (IleH2.4 in the universal nomenclature of Atchley et al 19) is spatially close to amino acids LeuH1.10, PheH1.7 and LysH2.1 of E47. Of these residues, it is unlikely that the I58V mutation will alter the interaction with LeuH1.10. This is because of the fact that, even though the substitution of isoleucine with valine results in the loss of a methyl group at this position, this side chain is on the opposite side of the amino acid to LeuH1.10 to which it is physically close. PheH1.7 is an important contact residue in both E47 homo- and heterodimerization complexes, forming part of the dimer interface. In NeuroD and HES7, the homologous residue is a leucine, and Longo et al18 predict that the smaller leucine residue packs less well than phenylalanine in the H1.7 position of E47, and produces a cavity across the dimer interface that may destabilise the NeuroD homodimer. The I58V mutation in HES7 may increase the size of this proposed cavity, changing the homo- and/or heterodimerization properties of the mutant protein. Alternatively, if I58V alters the position of the LysH2.1 residue (which is part of the DNA-binding interface), HES7 DNA-binding affinity may be altered. Therefore, although the I58V mutant Hes7 showed normal activity in all functional assays when expressed on its own, and in combination with wild-type or D186Y mutant Hes7, we cannot exclude the possibility that under in vivo conditions, this mutant has altered functional properties. In addition, as the two individuals in this family who are heterozygous for the D186Y mutation demonstrate no overt phenotype, it is clear that D186Y alone is not sufficient to cause SCD, and only the individuals carrying both alleles are affected. Surprisingly, in the N-box assay, when these two mutants are co-expressed, repression levels are indistinguishable from that of wild type. This might suggest that some function of HES7 other than N-box binding (such as heterodimerization with E47) is required for normal somitogenesis. Although we have not formally excluded the presence of another undetected mutation, either in HES7 or in a different gene, we conclude that these two new mutant alleles of HES7 in combination are likely to be causative of SCD in this family.
It is interesting to note that the two affected individuals in this family, in addition to the vertebral defects characteristic of SCD, both manifested a defect in the vertebral arteries in the neck. Although this was not seen in the previous patient carrying a different mutation in HES7,6 they were not investigated using magnetic resonance angiography. Although we cannot rule out that it is a secondary consequence of the segmentation phenotype in the cervical region, given that similar vascular defects have not been reported for any case of SCD carrying mutations in DLL3, MESP2 or LFNG, this finding may be indicative of a phenotype unique to HES7.
Finally, we believe that our discovery of two more mutations in HES7 from sequencing a small patient cohort (∼10 patients) indicates that HES7 is likely to be a more important contributor to SCD than either MESP2 or LFNG. For both of the latter two genes, only a single mutation has been reported from sequencing a worldwide cohort of SCD cases (>100), compared with ∼27 in DLL3. Therefore, our results have the important clinical implication that HES7 should be sequenced along with DLL3 as the first priority in all new cases of SCD.
References
Turnpenny PD, Alman B, Cornier AS et al: Abnormal vertebral segmentation and the notch signaling pathway in man. Dev Dyn 2007; 236: 1456–1474.
Bulman MP, Kusumi K, Frayling TM et al: Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat Genet 2000; 24: 438–441.
Sparrow DB, Clements M, Withington SL et al: Diverse requirements for Notch signalling in mammals. Int J Dev Biol 2002; 46: 365–374.
Whittock NV, Sparrow DB, Wouters MA et al: Mutated MESP2 causes spondylocostal dysostosis in humans. Am J Hum Genet 2004; 74: 1249–1254.
Sparrow DB, Chapman G, Wouters MA et al: Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet 2006; 78: 28–37.
Sparrow DB, Guillen-Navarro E, Fatkin D, Dunwoodie SL : Mutation of HAIRY-AND-ENHANCER-OF-SPLIT-7 in humans causes spondylocostal dysostosis. Hum Mol Genet 2008; 17: 3761–3766.
Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R : Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev 2001; 15: 2642–2647.
Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS : Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 2002; 129: 1795–1806.
Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL : Lunatic fringe is an essential mediator of somite segmentation and patterning. Nature 1998; 394: 377–381.
Forsberg H, Crozet F, Brown NA : Waves of mouse lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr Biol 1998; 8: 1027–1030.
Saga Y, Hata N, Koseki H, Taketo MM : Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev 1997; 11: 1827–1839.
Sparrow DB, Chapman G, Turnpenny PD, Dunwoodie SL : Disruption of the somitic molecular clock causes abnormal vertebral segmentation. Birth Defects Res C Embryo Today 2007; 81: 93–110.
Davis RL, Turner DL : Vertebrate hairy and enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 2001; 20: 8342–8357.
Fischer A, Gessler M : Delta-Notch—and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res 2007; 35: 4583–4596.
Bessho Y, Hirata H, Masamizu Y, Kageyama R : Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev 2003; 17: 1451–1456.
Collins JS, Schwartz CE : Detecting polymorphisms and mutations in candidate genes. Am J Hum Genet 2002; 71: 1251–1252.
Bessho Y, Miyoshi G, Sakata R, Kageyama R : Hes7: a bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 2001; 6: 175–185.
Longo A, Guanga GP, Rose RB : Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry 2008; 47: 218–229.
Atchley WR, Terhalle W, Dress A : Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol 1999; 48: 501–516.
Acknowledgements
We thank the family for their cooperation, and Wendy Chua for technical assistance. We are extremely grateful to Ryoichiro Kageyama and Diane Fatkin for providing reagents. This work was funded by National Health and Medical Research Council (NHMRC) Project Grants 404804 and 635500 (SLD and DBS), a Pfizer Foundation Australia Senior Research Fellowship (SLD) and a NHMRC Senior Research Fellowship (SLD). This research was approved by St Vincent's Hospital Human Research Ethics Committee (H05/057), Darlinghurst, Sydney, NSW 2010, Australia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Sparrow, D., Sillence, D., Wouters, M. et al. Two novel missense mutations in HAIRY-AND-ENHANCER-OF-SPLIT-7 in a family with spondylocostal dysostosis. Eur J Hum Genet 18, 674–679 (2010). https://doi.org/10.1038/ejhg.2009.241
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.241
Keywords
This article is cited by
-
Whole genome sequencing in cats, identifies new models for blindness in AIPL1 and somite segmentation in HES7
BMC Genomics (2016)
-
Whole Genome Sequencing Identifies a Missense Mutation in HES7 Associated with Short Tails in Asian Domestic Cats
Scientific Reports (2016)
-
Whole exome sequencing in an Indian family links Coats plus syndrome and dextrocardia with a homozygous novel CTC1 and a rare HES7 variation
BMC Medical Genetics (2015)
-
ptk7 mutant zebrafish models of congenital and idiopathic scoliosis implicate dysregulated Wnt signalling in disease
Nature Communications (2014)
-
The role of notch signaling in bone development and disease
Hormones (2014)