Abstract
Energy-coupling factor (ECF) transporters are a large family of ATP-binding cassette transporters recently identified in microorganisms. Responsible for micronutrient uptake from the environment, ECF transporters are modular transporters composed of a membrane substrate-binding component EcfS and an ECF module consisting of an integral membrane scaffold component EcfT and two cytoplasmic ATP binding/hydrolysis components EcfA/A'. ECF transporters are classified into groups I and II. Currently, the molecular understanding of group-I ECF transporters is very limited, partly due to a lack of transporter complex structural information. Here, we present structures and structure-based analyses of the group-I cobalt ECF transporter CbiMNQO, whose constituting subunits CbiM/CbiN, CbiQ, and CbiO correspond to the EcfS, EcfT, and EcfA components of group-II ECF transporters, respectively. Through reconstitution of different CbiMNQO subunits and determination of related ATPase and transporter activities, the substrate-binding subunit CbiM was found to stimulate CbiQO's basal ATPase activity. The structure of CbiMQO complex was determined in its inward-open conformation and that of CbiO in β, γ-methyleneadenosine 5′-triphosphate-bound closed conformation. Structure-based analyses revealed interactions between different components, substrate-gating function of the L1 loop of CbiM, and conformational changes of CbiO induced by ATP binding and product release within the CbiMNQO transporter complex. These findings enabled us to propose a working model of the CbiMNQO transporter, in which the transport process requires the rotation or toppling of both CbiQ and CbiM, and CbiN might function in coupling conformational changes between CbiQ and CbiM.
Similar content being viewed by others
Introduction
Powered by ATP hydrolysis, ATP-binding cassette (ABC) transporters comprise a large superfamily of protein complexes that transport a variety of substrates across cell membranes. On the basis of the transport direction, ABC transporters are categorized into importers and exporters, both of which are composed of two transmembrane domains, and two nucleotide-binding domains (NBDs). Importers, usually found in prokaryotes, contain a periplasmic solute-binding protein (SBP) for substrate capture. Through extensive studies, a two state rock-and-switch model has been suggested for ABC transporters1,2,3,4,5,6,7,8.
In recent years, a new family of ABC transporters, known as energy-coupling factor (ECF) transporters, have been identified in bacteria9,10,11,12. ECF transporters mediate micronutrient uptake and are composed of four components, two cytoplasmic ATPase components EcfA and EcfA', and two transmembrane components namely EcfT for energy coupling and EcfS for substrate recognition. The EcfT-EcfA-EcfA' components comprise the ECF module (Figure 1A). To date two groups of ECF transporters have been identified, with group-I ECF transporters being an EcfS protein associated with a dedicated ECF module and the encoding genes located within one operon. Group-II ECF transporters include several EcfS proteins; their coding genes are distributed throughout the genome and they can bind to a common ECF module12. In some group-I ECF transporters, the EcfS component is comprised of two proteins, M and N, which fuse together into one polypeptide MN in some species9. The most striking divergence that sets ECF transporters apart from canonical ABC importers lies in the SBPs, such that ECF transporters utilize integral membrane EcfS proteins for substrate binding, whereas canonical ABC transporters utilize periplasmic SBPs to capture substrates13,14,15,16.
Activity of CbiMNQO. (A) Conceptual diagram showing the constitution and differences between groups I and II ECF transporters. (B) Transporter activity of different subunit combinations of CbiMNQO transporter complex detected by ICP-MS. Vertical axis represents concentration of Co2+ or Ni2+ detected in E. coli. Data represent mean concentration (PPB) per OD600 ± SE. Riboflavin ECF transporter ECF-RibU was used as a control. (C) Purification of different subcomplexes of CbiMNQO. (D-E) ATPase activity of CbiMNQO, CbiMQO, CbiQO, and CbiO (blue, orange, gray, and yellow, respectively). Vertical axis represents free phosphate released (velocity in D and concentration in E). (F) ATPase activity of wild-type CbiMNQO (WT) and H2D mutant in response to Co2+ concentrations of blue, WT and orange, H2D mutant.
Extensive studies have been carried out to dissect the mechanism of ECF transporters, especially that of group-II transporters. The substrate-specific binding proteins of group-II ECF transporters, such as RibU, ThiT, BioY, and FolT, have very low sequence similarity, but they adopt similar six-helical-bundle 3D structures17,18,19,20. The high binding affinities of these EcfS proteins for their substrates10,21 are attributed to buried substrate-binding pockets within the middle of transmembrane helices near the periplasmic side. Residues constituting the substrate-binding pockets dictate the substrate specificity, and are highly conserved within a specific EcfS across different species22. This forms a basis for predicting the substrates for ECF transporters from different species.
The assembly of the EcfS component with the EcfAA'T module was not known until the inward-open structures of ECF transporters were captured23,24,25. These structures reveal that EcfA/A' components have two NBDs that form a dimer required for ATP hydrolysis, and EcfT component acts like a scaffold that connects EcfS component with EcfA/A' components, whereas EcfS components within the structures, without exception, adopt conformations parallel to membrane planes. These studies provided a snapshot toward elucidating scenarios of transport processes, and laid the foundation for a rotation/toppling model of group-II ECF transporters22,26,27.
Compared to group-II ECF transporters, limited progress has been made in the understanding of group-I ECF transporters9,11. The best known group-I ECF transporters are the widely distributed CbiMNQO and NikMNQO that uptake Co2+ and/or Ni2+, which are cofactors of a number of essential metabolic enzymes in bacteria28,29,30. CbiMNQO/NikMNQO are composed of M/N, Q, and O subunits, corresponding to the EcfS, EcfT, and EcfA components of group-II ECF transporters, respectively (Figure 1A). The structure of NikM2 in complex with Ni2+ has been reported31, and compared to group-II EcfS proteins, NikM2 contains an additional N-terminal transmembrane helix and an extended loop essential for substrate coordination. This study suggests that group-I ECF transporters possess different substrate-binding mechanisms from that of group-II ECF transporters. In addition, although the M subunit alone can bind substrate, the transporter activity of CbiMNQO/NikMNQO depends on the N subunit32,33. The molecular understanding of group-I ECF transporters remains elusive, in part due to the lack of structural information for the complete transporter complex. Here, structural and biochemical analyses of the CbiMNQO transporter complex are presented. Our results support a rotation/toppling model and shed new lights on the mechanism of group-I ECF transporters.
Results
Characterization of CbiMNQO transporter
The CbiMNQO transporter was characterized by expression of the transporter complex and combinations of different subunits in E. coli. Transporter activity was assayed using inductively coupled plasma mass spectrometry (ICP-MS) (Figure 1B). The results showed that the complete CbiMNQO transporter possessed high cobalt uptake activity, whereas a complex composed of CbiM and CbiN subunits exhibited low but significant activity. Removal of CbiN demolished cobalt uptake activity, consistent with a previous report32. CbiMNQO transporter exhibited very low nickel uptake activity, about 8% of that of cobalt, indicating that CbiMNQO is highly specific for cobalt.
By adding a 6× His tag to the CbiQ subunit, the transporter complex was purified using affinity chromatography. CbiM, CbiQ, and CbiO subunits were present in substantial amounts (1/1/2 molar ratio), but amounts of the CbiN subunit were reduced after each purification step (Figure 1C; Supplementary information, Figure S1). These results suggest that CbiM, CbiQ, and CbiO form a stable subcomplex (CbiMQO) and that the interaction of CbiN with the subcomplex is weak and dynamic. The formation of a stable CbiMQO subcomplex was verified when the three subunits were coexpressed and purified in the absence of CbiN (Figure 1C). Interestingly, the presence of CbiN appears to have very little effect on the ATPase activity of the complex. The ATP turnover rate (kcat) is 29.7 min−1 for CbiMNQO and 33.9 min−1 for CbiMQO (Km = 153.8 μM for CbiMNQO and Km = 150.7 μM for CbiMQO). This suggests that CbiN is not required for ATP binding and hydrolysis, at least in vitro (Figure 1D).
In addition to the above observations, CbiQ and CbiO subunits were found to form a stable subcomplex in the absence of CbiM in vitro (Figure 1C). This CbiQO complex shows only basal ATP hydrolysis activity (kcat = 2.5 min−1 and Km = 277.3 μM). However, the CbiO subunit alone is unstable and lacks ATPase activity (Figure 1D and 1E). These data suggest that CbiM is either required for or greatly stimulates the ATP hydrolysis by the CbiMNQO transporter. This stimulation is independent of the presence or binding of Co2+(Figure 1F). Alteration of the cobalt concentration or substitution of the cobalt-binding site on CbiM to abolish Co2+ binding/uptake activity, (using the H2D mutant as in Yu et al.31), has little impact on the ATPase activity of the transporter.
Overall transporter structure and subunit interactions
The underlying molecular mechanism of group-I cobalt ECF transporters was studied by purifying the CbiMNQO transporter complex for crystallization. Crystals were grown using the vapor diffusion method, and anisotropic X-ray diffraction data were collected. Complex structure determination was aided by separately crystallizing a CbiO subunit containing an E166Q substitution that was more stable than the wild type. The determined structure was then used as a search model to solve the transporter complex structure (Supplementary information, Figure S2). The structure was refined to 2.8 Å with R and Rfree of 21.4% and 24.9%, respectively. The final model contains residues 1-207 of CbiM, residues 1-244 of CbiQ, and residues 1-280 of CbiO, but no CbiN. This new model was then used to solve a 3.5 Å resolution data set collected from crystals grown in lipid cubic phase (LCP). This structure, obtained using crystals grown in a lipid bilayer environment, also did not show the presence of CbiN. Therefore, the structure obtained was concluded to represent the CbiMQO subcomplex (Supplementary information, Table S1). The overall structure obtained using the lipid-cubic-phase method is very similar to that obtained using the vapor diffusion method, both showing an inward-open conformation of the complex (r.m.s.d. = 1.6 Å). Therefore, the 2.8 Å CbiMQO structure obtained using the vapor diffusion method was used for further description and analysis.
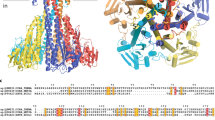
The CbiMQO complex is composed of one CbiM subunit, one CbiQ subunit, and two CbiO subunits, which confirms a stoichiometry of 1:1:2. The overall structure is similar to that of group-II ECF transporters and forms a double-cone, with the transmembrane subunits CbiM and CbiQ forming the top half of the cone and two cytoplasmic CbiO subunits forming the bottom half. The two CbiO proteins, both nucleotide-free, form a homodimer mainly through their C-terminal domains (Figure 2A). Similar to group-II EcfS proteins, CbiM lies horizontally along the lipid membrane and its transmembrane helices (SM0-6) are roughly parallel to the membrane surface. Thus, the substrate-binding site is open to the cytoplasmic side of the membrane through a large cleft formed between CbiO and CbiQ (Figure 2A and 2B). The unique feature of CbiM, compared with group-II EcfS is the presence of an additional helix at its N terminus that packs with the backsides of CbiQ and CbiM (Figure 2C). For comparison with group-II EcfS proteins, the transmembrane helix at the very N terminus was numbered SM0 (colored red), which is followed by SM1-6 and preceded by the N terminus loop L0 (Figure 3A).
Structure of CbiMQO complex. (A-C) Overall structure of CbiMQO viewed parallel to the membrane. CbiO dimer is shown with a light-blue surface model. CbiM is shown as a surface model in A and a ribbon model in B and C with SM1-5 in gold and SM0/L0 loop in red. CbiQ is shown as a ribbon model with TM1-5 in yellow and CH2-3 in magenta.
Interactions between different CbiMQO complex subunits. (A) Ribbon diagram of CbiM structure. Color codes are the same as in Figure 2. (B) Interactions between transmembrane helices of CbiQ and CbiM. Residues from CbiQ (light-blue sticks) forming hydrophobic interactions with those from CbiM (orange) are shown. Residue Phe75 from TM4 is shown as a spherical model. (C) Hydrophobic interaction surface formed between CH2-3 of CbiQ and SM1, 3, and 6 of CbiM. CbiQ is shown with electrostatic potential surface. CbiM is shown as a ribbon model and residues Ala46 and Ala50 from SM1 are shown in blue. (D) Interactions between CH2-3 of CbiQ and CbiO dimer. CbiO is shown with electrostatic potential surface. CH2-3 of CbiQ are shown as ribbons with residues involved shown with sticks. Arg169 and Arg207 from the XRX motifs are shown as spherical model models. (E) Cobalt transporter activity of CbiMNQO containing interface mutations. Riboflavin ECF transporter ECF-RibU was used as a control.
CbiQ is composed of five transmembrane helices (TM1-5) and four cytoplasmic helices (coupling helices 1-3/CH1-3 and the N terminal short helix), forming a C-shape surrounding the CbiM subunit (Figures 2B and 3B). Two of the cytoplasmic helices, coupling helix 2 and 3 (CH2 and CH3), form an “X” shape and separate CbiM from interaction with CbiO. CH2 and CH3 are followed, respectively, by R1 and R2 loops, which feature conserved XRX motifs24,34.
CbiQ functions like a scaffold connecting CbiM and CbiO, with extensive interactions between CbiQ and CbiM, and between CbiQ and CbiO. The interactions between CbiM and CbiQ are primarily hydrophobic, burying a total surface area of ∼6 014 Å2. There are two separate interaction surfaces between CbiQ and CbiM. One is composed of CbiM L0, L3, and L5 loops, which form hydrophobic interactions with a number of aliphatic residues from TM2, TM3, and TM4 of CbiQ (Figure 3B). It is notable that residue Phe75 from TM4 docks its side chain into the possible substrate-binding site on CbiM (Figure 3B; Supplementary information, Figure S3). This residue is conserved among cobalt/nickel transporters, but not other ECF transporters (Supplementary information, Figure S4). Mutation of Phe75 to Ala reduces the transporter activity to a large extent (Figure 3E), indicating an important role of this conserved Phe residue in cobalt/nickel-specific ECF transporters. This interaction surface buries 2 266 Å2 versus 1 756 Å2 in group-II LbECF-FolT, suggesting that the binding of group-I CbiM with the ECF module is much stronger than that of group-II EcfSs. The latter were found ready to dissociate from the transporter complex in the presence of ATP and substrate24,25. This might explain the specific recognition between CbiM and the ECF module in group-I transporter. The other interaction surface between CbiQ and CbiM is composed of CbiM SM1, SM3, and SM6, which forms a hydrophobic surface and a groove similar to that observed in group-II EcfS proteins25, and stack together with CH2 and CH3 of CbiQ. Two small side chain residues, Ala46 and Ala50, are present in SM1 (Figure 3C). Mutation of these two residues to Trp impairs transporter activity (Figure 3E).
The interface between CbiQ and CbiO involves both hydrophilic and hydrophobic interactions (Figure 3D). CH2 and CH3 of CbiQ bind into a deep groove formed by the helical and RecA subdomains of CbiO, mediating major interactions and energy coupling with CbiO. The strictly conserved residues Arg169 and Arg207 from the R1/R2 loops (XRX motifs) bind directly with residue Asp102 of CbiO (Supplementary information, Figures S4, S5A, and S5B). In addition, CH2 and CH3 harbor several conserved arginine residues, Arg150, Arg196, and Arg192, which form hydrogen bonds with residues Asp91 and Asp92 in CbiO (Supplementary information, Figures S4, S5C, and S5D). There are also a number of hydrophobic residues in CH2 and CH3 that form van der Waals' interactions with residues in CbiO (Figure 3D). Using the ICP-MS assay, mutations of these charged residues were found to significantly reduce transporter activity (Figure 3E), suggesting important roles for these residues in coupling energy and conformational changes between CbiO and CbiQ.
L1 loop gates substrate binding and release
As a representative of group-I EcfS proteins, CbiM is distinct from group-II EcfS proteins by possessing an extra transmembrane helix, SM0, which was flanked by the L0 loop at the CbiM N-terminus (Figure 3A). Such a structure in NikM2 has been reported to be responsible for substrate binding31. CbiM structure in the present CbiMQO complex is captured in a substrate-free conformation, and superimposition of CbiM with the substrate-bound NikM2 structure yields a root mean square deviation of 2.0 Å (Figure 4A). Comparison of these two structures reveals significant conformational difference in their L1 loops. In NikM2, the L1 loop covers the top of the nickel-binding site and adopts a “close” conformation. In addition, the Nɛ2 atom of L1 loop residue His67 coordinates the Ni2+ ion with the nitrogen atom of the free N-terminal amino group of residue Met1 as well as the main chain amide nitrogen and Nδ1 atom of residue His2, yielding a square planar substrate geometry. In the present substrate-free CbiM structure, L1 loop is displaced from the substrate-binding site to adopt an “open” conformation. Thus, residue His69 (corresponding to His67 in NikM2) is far away from residues His2 and Met1, ruining the substrate-binding site. The importance of these residues was verified using transporter assay, in which the results showed that mutation of residue His2 or His69 to Asp abolished the transporter activity. As a control, mutation of residue His102 from the L3 loop to Asp exhibited a mild effect on transporter activity (Figure 4B). Further examination of sequence alignment showed that residues in CbiM corresponding to those forming hydrogen-bonding networks to stabilize the substrate-binding site in NikM2 are quite different (Supplementary information, Figure S6). It is possible that the interaction network and the bonding strength in the substrate-binding sites of NikM2 and CbiM are different, which may be attributed to their distinct substrate specificity.
Structural comparison of CbiM with NikM2. (A) Superimposition of the structures of substrate-free CbiM (gold) with substrate-bound NikM2 (gray, PDB code: 4M58) shows conformational differences. L1 loops of CbiM and NikM2 are in red and blue, respectively, and residues coordinating Ni2+ (blue sphere) in NikM2 and corresponding residues in CbiM are shown with sticks. (B) Cobalt transporter activity of CbiMNQO containing substrate-binding site mutations. Riboflavin ECF transporter ECF-RibU was used as a control.
Another significant conformational difference between the substrate-free CbiM and substrate-bound NikM2 was detected in their L5 loops. Compared to NikM2, there is a seven-residue deletion in the L5 loop of CbiM (Supplementary information, Figure S6). In NikM2, the L5 loop forms hydrophobic interactions with the L0 and L1 loops, and aids in stabilizing the substrate-binding conformation. In CbiM, there are hydrophobic interactions between the L5 loop and the L0 loop, and the L5 loop forms additional interactions with the β1-β2 hairpin loop connecting TM3-TM4 of CbiQ (Figure 3B). Taken together, these data suggest that the L1 loop plays a critical role in the gating of substrate binding and release in group-I ECF transporters, and the L5 loop may also be involved in this process. The conformational differences observed between the substrate-free CbiM and substrate-bound NikM2 may reflect the conformational changes during the substrate binding and release process. It is noteworthy that similar conformational changes in the L1 loop of group-II EcfS proteins have been observed20, suggesting that both group-I and II ECF transporters use a similar mechanism to control the substrate binding and release process, even though they have evolved distinct structural features.
Conformational changes of CbiO upon ATP binding and product release
In the process of solving the CbiMQO structure, a 1.45 Å resolution structure of the CbiO dimer (containing an E166Q mutation) was also determined in complex with ATP analogue β, γ-methyleneadenosine 5′-triphosphate (AMPPCP) and Mg2+ (CbiO-ACP) (Figure 5A). Typical motifs involved in ATP binding and hydrolysis were identified including Walker-A (WA), Walker-B (WB), LSGGQ loop, His switch, and Q-loop. AMPPCP binds in the cleft of the CbiO dimer interface formed by the RecA-like subdomain and a neighboring helical subdomain. Specifically, the adenine base is stabilized by the highly conserved residue Phe14 from one subunit and the LSGGQ main chain from another subunit (LSGGQ'). In addition, the ATP's ribose forms four hydrogen bonds, directly or via water (Wat12 and Wat104), with residues Lys19, Arg136′, and Gln145′. Meanwhile, the α, β, and γ-phosphates form at least 18 hydrogen bonds with residues from WA, WB (through Wat64), LSGGQ' loop, His switch, and Q-loop (Figure 5B).
Conformational change of CbiO upon ATP binding and product release. (A) Surface model of CbiO-ACP structure. RecA-like subdomain is in light orange and helical subdomain in gold. Walker A, Walker B, Q loop, LSGGQ loop, and H-switch are in red, green, blue, magenta, and purple, respectively. AMPPCP is shown with a stick model and Mg2+as a sphere. (B) Schematic illustrations show the interactions between CbiO and AMPPCP. Hydrogen bonds are shown with dashed lines and hydrophobic interactions are shown with triple-dashed lines. (C) Superimposition of the structures of CbiO-open (light blue) and CbiO-ACP (color codes are the same as in A) showing the conformational changes induced by ATP binding and product release. (D) Zoom in view of conformational changes of Walker A and LSGGQ loop regions shown in C. (E) Zoom in view of conformational changes of Q loop shown in C. (F) A simplified model showing conformational change of CbiO after ATP binding. Helical subdomains and RecA-like subdomains are represented by blue ovals and rectangles, respectively. ATP molecules are shown with red stars.
In contrast to the nucleotide-free, open conformation of CbiO in the CbiMQO complex (CbiO-open), CbiO-ACP is captured in a closed conformation (Figure 5C), which may resemble the ATP-binding active state of the intact transporter complex and, thus, allows us to pinpoint the ATP-binding and product-releasing induced conformational changes during the transport process. Compared to the CbiO-open structure, the two subunits in CbiO-ACP moved toward each other, with significant movement and/or conformational changes taking place in WA, the LSGGQ loop, and the Q loop (Figure 5C, 5D, and 5E). Eventually forming a closed dimer, the WA motif from one CbiO subunit and the LSGGQ motif from the other form a sandwich with ATP in the middle (Supplementary information, Movie S1). Considering the helical subdomain and RecA-like subdomain of each subunit as two independent rigid bodies, movement induced by ATP binding to the CbiO dimer can be simplified as rigid body rotations (Figure 5F). Upon ATP binding, the helical subdomain rotates around a vertical axis toward the ATP binding site, whereas the RecA-like subdomain rotates around a horizontal axis to the center of the CbiO dimer. After ATP hydrolysis, this movement is reversed to release the final products, ADP and phosphate. The CbiO dimer interacts directly with the “X”-shaped coupling helices CH2-CH3 of CbiQ, via conserved hydrophilic and hydrophobic interactions (Figures 2 and 3D). As a result, the conformational changes in CbiO are transferred to CbiQ and then further to CbiM, driving cross-membrane substrate transport.
Discussion
In this study, we reconstituted and characterized different component combinations of the CbiMNQO cobalt transporter complex. Complex ATPase activity was found to be greatly stimulated by the substrate-binding subunit CbiM. This stimulation is independent of the presence of substrate, and is different from the most studied canonical ABC transporters, whose ATPase activity largely depends on the presence of substrate-binding SBPs35,36,37. Interestingly, the CbiN subunit, which is essential for cobalt transport, was found not required for the complex ATPase activity. The structures of CbiMQO and CbiO-ACP complexes were solved and then used to analyze the underlying molecular mechanism of the CbiMNQO transporter by: (1) revealing the interactions among reconstituted subunits and verifying those interactions using a transporter assay system, (2) identifying the essential role of the CbiM L1 loop in substrate gating that resembles what has been seen in the EcfS proteins of group-II ECF transporters, and (3) illustrating the conformational changes within the CbiO dimer that are induced by ATP binding and product release.
As the first complex structure of a group-I ECF transporter, CbiMQO exhibits several features that are distinct from reported structures of group-II ECF transporters. Superimposition of the CbiMQO structure with the LbECF-FolT structure reveals significant differences (r.m.s.d. = 3.0 Å). One of the most striking differences is the substrate-binding proteins CbiM and FolT (Figure 6A, right). They possess different numbers of transmembrane helices (seven and six, respectively), and the position and orientation of each respective helix in CbiM and FolT differ significantly. These observations explain the unique substrate-binding mode of CbiM/NikM. Another difference is observed in the transmembrane helices TM1-5 of CbiQ, which appears to swing toward the substrate-binding subunit in CbiMQO unlike those in LbECF-FolT (Figure 6A, left). A similar conformational difference has been identified for a single EcfT protein in different group-II ECF transporters from the same species25. These structural observations imply that TM1-5 of CbiQ/EcfT may undergo a swing-like movement during transport. This movement and the movement of the CH2-CH3 helices of CbiQ/EcfT, may need to be coupled to the substrate-binding proteins CbiM/EcfS to load and unload substrate during the transport process. As CbiM features an extra N-terminal SM0 helix and L0 loop that are required for substrate binding, the coupling process in CbiMNQO may require CbiN, which is essential for transporter activity. Considering the results that CbiM alone can stimulate the ATPase activity of CbiQO and that CbiN is dynamically integrated into the CbiMNQO complex in vitro, we postulate that CbiN may interact with CbiM and CbiQ, and that CbiN may be required for coupling TMs movements of CbiQ to CbiM (i.e., CbiN may be required for resetting the substrate-binding site in CbiM). On the basis of the above analysis, we modeled CbiN, which is predicted to contain two transmembrane helices, around the SM0 region of CbiM (Figure 6B).
Structural differences between CbiMNQO and LbECF-FolT. (A) Superimposition of CbiMQO with LbECF-FolT (PDB code: 4HUQ) showing differences between CbiQ(yellow)/EcfT(green) and CbiM(gold)/FolT(light blue) (left and right, respectively). (B) Possible location of CbiN in CbiMNQO transporter. CbiN is shown as a ribbon cartoon in blue.
We summarize the above hypothesis in a simplified two-state working model in which CbiMNQO exists mainly in two conformational states during the transport process, the outward-facing and inward-facing states (Figure 7). In the outward-facing state, the CbiO dimer binds ATP and adopts a closed conformation. Accordingly, CbiQ and CbiM adopt upright conformations that allow the L1 and L0 loops of CbiM to coordinate cobalt in the substrate-binding site, and CbiN bridges CbiM and CbiQ from the backside. ATP hydrolysis and subsequent product release powers the transition from the outward-facing state to inward-facing state, in which the CbiO dimer shifts to open conformation. This conformational change leads to the movement of CH2-CH3 helices and TM1-5 of CbiQ, which are coupled to CbiM through the CbiM hydrophobic surface groove and CbiN, respectively. As a result, CbiM rotates or topples within the membrane, releasing the substrate into the cytoplasm and completing the transport process. ATP binding pushes CbiQ and CbiM back to upright positions and resets the complex to an outward-facing conformation, ready to start another transport cycle. Loss of CbiN will disrupt the proper conformational transmission between TM1-5 of CbiQ and CbiM, but not the ATPase activity of the complex, thereby uncoupling ATP hydrolysis with cobalt transport (Supplementary information, Figure S7). Our data suggest that the group-I cobalt ECF transporter CbiMNQO uses a rotation or toppling mechanism similar to what has been proposed for group-II ECF transporters22,23,24,26,27, but features a different substrate-binding mode and a different energy-coupling process.
Probable working model of CbiMNQO transporter. CbiQ is in yellow, and TM1-5 and CH2-3 are indicated with a rectangle and two crossed cylinders, respectively. CbiM is shown with cylinders in light blue, except for L0 andL1 loops, and SM0 in red. CbiN is shown as a surface model in blue. CbiO dimer is shown as a cartoon in gray.
Materials and Methods
Protein expression and purification
Genes encoding the four subunits of the cobalt ECF transporter, CbiM, CbiN, CbiQ, and CbiO were amplified by PCR from Rhodobacter capsulatus genomic DNA. The gene ID and predicted molecular mass are: CbiM, GI: 294476393, 22.5 kDa; CbiN, GI: 294476392, 10.6 kDa; CbiQ, GI: 294476391, 25.9 kDa; CbiO, GI: 294476390, 29.4 kDa. Genes were cloned into pETDuet and pRSFDuet vectors to obtain two expression plasmids: pETDuet-CbiM-CbiN and pRSFDuet-CbiQ-CbiO. A tag of six histidine residues was added at the N terminus of CbiQ. To identify the expression of CbiN, a flag tag was added at the N terminus of CbiN. For purification of CbiMQO subcomplex, CbiM was cloned into pETDuet to get pETDuet-CbiM. For purification of CbiO, CbiO gene was cloned into pETDuet vector with a 6× His or MBP tag at the N terminus. CbiO E166Q mutation and mutations of CbiMNQO for transport assay and ATPase activity assay were constructed using one-step PCR and verified by sequencing.
For CbiMNQO protein expression, pETDuet-CbiM-CbiN and pRSFDuet-CbiQ-CbiO plasmids were co-transformed into E. coli BL21 (DE3) and cultured at 37 °C using lysogeny broth (LB) medium. Protein was induced by 0.25 mM β-D-thiogalactopyranoside (IPTG) at A600 of about 1.0. After 12 h induction, the cells were collected, and homogenized in buffer A containing 50 mM Tris-HCl, pH 8.0 and 300 mM NaCl, and lysed using French Press. Cell debris was removed by centrifugation. The supernatant was collected and applied to ultracentrifugation at 150 000× g for 1 h. Membrane fraction was incubated with 1% (w/v) n-dodecyl-β-D-maltopyranoside (DDM, Bluepus) for 2 h at 4 °C. After another centrifugation step at 20 000× g for 1 h, the supernatant was loaded onto a Ni2+-NTA affinity column (Qiagen) for purification. Protein sample was eluted with buffer A plus 250 mM imidazole and 0.018% DDM, and further purified by gel filtration, in which buffer B containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 0.18% NG (Anatrace) was used. The peak fraction was collected and concentrated to ∼ 10 mg/ml for crystallization. A similar protocol was used in the expression and purification of subcomplexes CbiMQO, CbiQO, and CbiMNQO with flag tag. For the Selenium-methionine CbiMNQO complex expression, a published protocol was adopted38, and the purification also followed the above steps.
For CbiO expression, pETDuet-CbiO (wild type or E166Q) was transformed into E. coli BL21 (DE3) strain. The transformed cells were grown at 37 °C and induced by 0.25 mM IPTG for 12 h at 25 °C. The cells were harvested, resuspended in buffer C (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1 mM ATP, 1mM EDTA) and lysed using French Press. The total lysate was centrifuged at 20 000× g for 45 min before the supernatant was loaded onto a Ni2+-NTA column for purification. After competitive wash with buffer C plus 25 mM imidazole, protein sample was eluted with buffer C plus 250 mM imidazole, and further purified by gel filtration. Peak fractions were collected and concentrated for subsequent studies.
Crystallization, data collection and structure determination
Crystallization of CbiO containing E166Q mutation was carried out by sitting drop vapor diffusion method at 4 °C. Prior to crystallization, 2 mM β, γ-methyleneadenosine 5′-triphosphate (AMPPCP) and MgCl2 were added to the protein sample (8 mg/ml). The best crystals were grown under a condition containing 0.64 M sodium acetate, pH 4.6, 18% PEG 3350 (protein: reservoir volume in 1:1 ratio). Crystals used for data collection were directly flash frozen in a liquid nitrogen stream at 100 K. A 1.45 Å resolution data set was collected at the BL19U1 beamline at the Shanghai Synchrotron Radiation Facility (SSRF). The crystals belong to the space group P212121 with unit cell dimensions of a = 71.3 Å, b = 74.4 Å, c = 106.8 Å.
Crystallization of CbiMNQO complex was carried out using both vapor diffusion method and LCP method. For crystallization with vapor diffusion method, protein was concentrated to 8 mg/ml. The best crystals were grown under 1 M ammonium sulfate, 0.1 M ADA, pH 6.5 condition, and were directly flash frozen in liquid nitrogen for data collection (the Selenium-methionine crystals were also obtained using this method). To avoid reflection overlap in data collection, the longest c axis is aligned as close as possible to the spindle axis of crystal rotation. After numerous synchrotron trips, the best native data set and Se-SAD data set were collected at BL19U1 beamline at SSRF to 2.8 and 3.6 Å resolution, respectively. For LCP crystallization, protein was concentrated to 15 mg /ml and mixed with 1.5-folds of monolein in a coupled syringe device to make LCP. Protein containing LCP was dispersed onto a glass plate as 50 nl bolus, and covered with 800 nl of precipitant solution using a Gryphon robot. Rod-shaped crystals were grown in precipitant solution containing 0.1 M NaCl, 35-48% PEG 400, 0.1 M MES, PH 6.5 at 20 °C. A 3.5 Å resolution data set was collected at BL18U beamline. The crystals grown under vapor diffusion method belong to the space group C2221 with unit cell dimensions of a = 63.7 Å, b = 220.6 Å, c = 301.3 Å, whereas the crystals grown using LCP method belong to the space group P212121 with unit cell dimensions of a = 96.3 Å, b = 134.9 Å, c = 206.8 Å. All the data sets were processed with HKL300039 and XDS40.
Owing to the pathology of pseudo-translational symmetry in the crystals of CbiMQO, traditional phasing methods in crystallography failed for structure determination. Therefore, CbiO (E166Q) dimer complexed with AMPPCP was first crystallized and the structure was solved using molecular replacement with the group-II EcfA-EcfA' structure (PDB ID: 4HUQ) as the search model41. CbiO-ACP structure was then rotated and translated into the lattice of CbiMQO crystals to provide the initial phases. The phases were then improved using multi-crystal averaging42 including CbiMQO native data, the non-isomorphous CbiMQO selenium-substituted SAD data, and the CbiMQO in LCP data. The crude structural model was built in COOT manually43, and finally refined with phenix44. The sequence was validated with the anomalous difference map of CbiMQO Se-SAD data. Data collection and structural refinement statistics were summarized in Supplementary information, Table S1.
Model of CbiN
The 3D model of CbiN was predicted using CPHmodels server (http://www.cbs.dtu.dk/services/CPHmodels/)45. Then the model was modified to delete the loops and keep the two transmembrane helixes alone. On the basis of our experimental results and reported facts, the CbiN model was placed in COOT near the interface between CbiM and CbiQ.
Co2+ transport assay
To measure the cobalt transporter activity, the corresponding genes encoding the transporter complex subunits were transformed into BL21 (DE3) and were cultured in 5 ml LB medium at 37 °C overnight. Then the cells were inoculated to 50 mL LB medium containing 0.01 mM IPTG and 1 μM CoCl2. After growth at 37 °C for 12 h, the cells were collected and washed three times successively with solutions containing 1 mM EDTA, 100 mM NaCl, and ultrapure water (18.2 MΩ·cm). Then the samples were digested with 1 ml concentrated nitric acid (Baker Instra-Analyzed; Avantor Performance Materials) and diluted to 10.0 ml with ultrapure water. The internal standard Indium was added to the acid prior to digestion for monitoring technical errors and plasma stability in the ICP-MS instrument. After samples and controls were prepared, 59Co analysis was performed with an ICP-MS (NexION 300D; PerkinElmer) coupled to Apex desolvation system and SC-4 DX autosampler (Elemental Scientific, Omaha, NE, USA). All samples were normalized at a wavelength of 600 nm (A600). Tests were repeated at least three times independently.
ATPase activity assay
The ATPase activity was determined following a previously published protocol with minor modifications46. Briefly, the reaction was initiated by adding 0.5 mM ATP into a 20 μl mixture containing 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.018% DDM, 5% (v/v) glycerol, 0.5 mM MgCl2, and 0.5 μM protein (1 μM for CbiO). The reaction was carried out at 37 °C for a specified time, and was stopped by the addition of 200 μl dye buffer (a mixture of 0.045% (w/v) malachite green and 4.2% (w/v) ammonium molybdate tetrahydrate in 4 N HCl in a 1:3 ratio) followed by the addition of 25 μl 34% (w/v) sodium citrate solution after 1 min. For Km/kcat detection, ATP was added at a series concentrations and the reaction time for CbiMNQO, CbiNQO, and CbiQO were 5, 5, and 10 min, respectively. Absorbance was then measured at 660 nm. The amount of inorganic phosphate released was calculated as the standard curve established by known amounts of NaH2PO4.
Accession codes
The atomic coordinates and structure factors for the structures have been deposited in the Protein Data Bank (PDB) with accession codes 5X3X, 5X40, and 5X41.
Author Contributions
ZB and XQ designed and performed the majority of the experiments; SH and KX contributed to protein expression and crystallization; FH and MZ contributed to protein expression and assay analysis; JC and DC contributed to transport assay analysis; WZ and DL carried out the LCP crystallization. JW, ZB, and PZ solved the structures. PZ conceived the project and wrote the manuscript with inputs from all authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
Locher KP, Lee AT, Rees DC . The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 2002; 296:1091–1098.
Hvorup RN, Goetz BA, Niederer M, Hollenstein K, Perozo E, Locher KP . Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science 2007; 317:1387–1390.
Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J . Crystal structure of a catalytic intermediate of the maltose transporter. Nature 2007; 450:515–521.
Oldham ML, Chen J . Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science 2011; 332:1202–1205.
Khare D, Oldham ML, Orelle C, Davidson AL, Chen J . Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell 2009; 33:528–536.
Hollenstein K, Frei DC, Locher KP . Structure of an ABC transporter in complex with its binding protein. Nature 2007; 446:213–216.
Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC . The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science 2008; 321:250–253.
Gerber S, Comellas-Bigler M, Goetz BA, Locher KP . Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science 2008; 321:246–250.
Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T . Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J Bacteriol 2006; 188:317–327.
Duurkens RH, Tol MB, Geertsma ER, Permentier HP, Slotboom DJ . Flavin binding to the high affinity riboflavin transporter RibU. J Biol Chem 2007; 282:10380–10386.
Hebbeln P, Rodionov DA, Alfandega A, Eitinger T . Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci USA 2007; 104:2909–2914.
Rodionov DA, Hebbeln P, Eudes A, et al. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 2009; 191:42–51.
Rees DC, Johnson E, Lewinson O . ABC transporters: the power to change. Nat Rev Mol Cell Biol 2009; 10:218–227.
Chen J . Molecular mechanism of the Escherichia coli maltose transporter. Curr Opin Struct Biol 2013; 23:492–498.
Zolnerciks JK, Andress EJ, Nicolaou M, Linton KJ . Structure of ABC transporters. Essays Biochem 2011; 50:43–61.
Locher KP . Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol 2016; 23:487–493.
Zhang P, Wang J, Shi Y . Structure and mechanism of the S component of a bacterial ECF transporter. Nature 2010; 468:717–720.
Erkens GB, Berntsson RP, Fulyani F, et al. The structural basis of modularity in ECF-type ABC transporters. Nat Struct Mol Biol 2011; 18:755–760.
Berntsson RP, ter Beek J, Majsnerowska M, et al. Structural divergence of paralogous S components from ECF-type ABC transporters. Proc Natl Acad Sci USA 2012; 109:13990–13995.
Zhao Q, Wang C, Wang C, et al. Structures of FolT in substrate-bound and substrate-released conformations reveal a gating mechanism for ECF transporters. Nat Commun 2015; 6:7661.
Eudes A, Erkens GB, Slotboom DJ, Rodionov DA, Naponelli V, Hanson AD . Identification of genes encoding the folate- and thiamine-binding membrane proteins in Firmicutes. J Bacteriol 2008; 190:7591–7594.
Zhang P . Structure and mechanism of energy-coupling factor transporters. Trends Microbiol 2013; 21:652–659.
Wang T, Fu G, Pan X, et al. Structure of a bacterial energy-coupling factor transporter. Nature 2013; 497:272–276.
Xu K, Zhang M, Zhao Q, et al. Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis. Nature 2013; 497:268–271.
Zhang M, Bao Z, Zhao Q, et al. Structure of a pantothenate transporter and implications for ECF module sharing and energy coupling of group II ECF transporters. Proc Natl Acad Sci USA 2014; 111:18560–18565.
Slotboom DJ . Structural and mechanistic insights into prokaryotic energy-coupling factor transporters. Nat Rev Microbiol 2014; 12:79–87.
Finkenwirth F, Sippach M, Landmesser H, et al. ATP-dependent conformational changes trigger substrate capture and release by an ECF-type biotin transporter. J Biol Chem 2015; 290:16929–16942.
Okamoto S, Eltis LD . The biological occurrence and trafficking of cobalt. Metallomics 2011; 3:963–970.
Lebrette H, Brochier-Armanet C, Zambelli B, et al. Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins. Structure 2014; 22:1421–1432.
Desguin B, Goffin P, Viaene E, et al. Lactate racemase is a nickel-dependent enzyme activated by a widespread maturation system. Nat Commun 2014; 5:3615.
Yu Y, Zhou M, Kirsch F, et al. Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters. Cell Res 2013; 24:267–277.
Siche S, Neubauer O, Hebbeln P, Eitinger T . A bipartite S unit of an ECF-type cobalt transporter. Res Microbiol 2010; 161:824–829.
Kirsch F, Eitinger T . Transport of nickel and cobalt ions into bacterial cells by S components of ECF transporters. Biometals 2014; 27:653–660.
Neubauer O, Alfandega A, Schoknecht J, Sternberg U, Pohlmann A, Eitinger T . Two essential arginine residues in the T components of energy-coupling factor transporters. J Bacteriol 2009; 191:6482–6488.
Orelle C, Ayvaz T, Everly RM, Klug CS, Davidson AL . Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc Natl Acad Sci USA 2008; 105:12837–12842.
Davidson AL, Shuman HA, Nikaido H . Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc Natl Acad Sci USA 1992; 89:2360–2364.
Liu CE, Liu PQ, Ames GF . Characterization of the adenosine triphosphatase activity of the periplasmic histidine permease, a traffic ATPase (ABC transporter). J Biol Chem 1997; 272:21883–21891.
Qi X, Lin W, Ma M, et al. Structural basis of rifampin inactivation by rifampin phosphotransferase. Proc Natl Acad Sci USA 2016; 113:3803–3808.
Minor W, Cymborowski M, Otwinowski Z, Chruszcz M . HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 2006; 62:859–866.
Kabsch W . Xds. Acta Crystallogr D Biol Crystallogr 2010; 66:125–132.
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ . Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–674.
Cowtan K . Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr 2010; 66:470–478.
Emsley P, Cowtan K . Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132.
Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213–221.
Nielsen M, Lundegaard C, Lund O, Petersen TN . CPHmodels-3.0--remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res 2010; 38:W576–581.
Tang WK, Xia D . Role of the D1-D2 linker of human VCP/p97 in the asymmetry and ATPase activity of the D1-domain. Sci Rep 2016; 6:20037.
Acknowledgements
We thank the staff members at BL19U/18U of National Center for Protein Science Shanghai (NCPSS) and BL17U of Shanghai Synchrotron Radiation Facility (SSRF) for technical assistance in data collection, and the staff at the core facility center of Institute of Plant Physiology and Ecology for X-ray diffraction experimental test and analysis. We thank Dr David Degen from Rutgers University for manuscript reading and revision. This work was supported by grants from the Ministry of Science and Technology of China (2015CB910900 to PZ and 2015CB910104 to JW), the National Natural Science Foundation of China (31670755 and 31322016 to PZ), the Chinese Academy of Sciences (QYZDB-SSW-SMC006 to PZ and 2015LH0027 to FH), and the Shanghai Municipal Science and Technology Commission. Research in DL Lab is supported by the 1000 Young Talent Program, the Shanghai “Pujiang Talent” Program (15PJ1409400), and the National Natural Science Foundation of China (31570748).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
SDS-PAGE and western blot results of the samples during the purification process. (PDF 275 kb)
Supplementary information, Figure S2
Representative 2FoFc electron density contoured at 1.5σ level. (PDF 307 kb)
Supplementary information, Figure S3
Residue Phe75 from TM4 docks its side chain into the empty substrate binding site of CbiM (PDF 121 kb)
Supplementary information, Figure S4
Sequence alignment of CbiQ, NikQ and group-II EcfT from different species. (PDF 464 kb)
Supplementary information, Figure S5
Hydrogen-bonding Interactions between CH2-CH3 of CbiQ and CbiO. (PDF 448 kb)
Supplementary information, Figure S6
Sequence alignment of CbiM and NikM. (PDF 477 kb)
Supplementary information, Figure S7
A model shows the uncoupling of ATPase activity with cobalt transport caused by loss of CbiN. (PDF 113 kb)
Supplementary information, Table S1
Statistics of data collection and structure refinement. (PDF 108 kb)
Rights and permissions
About this article
Cite this article
Bao, Z., Qi, X., Hong, S. et al. Structure and mechanism of a group-I cobalt energy coupling factor transporter. Cell Res 27, 675–687 (2017). https://doi.org/10.1038/cr.2017.38
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2017.38
Keywords
This article is cited by
-
Cryo-EM structure and molecular mechanism of abscisic acid transporter ABCG25
Nature Plants (2023)
-
Identification of inhibitors targeting the energy-coupling factor (ECF) transporters
Communications Biology (2023)
-
Expulsion mechanism of the substrate-translocating subunit in ECF transporters
Nature Communications (2023)
-
Beneficial elements: New Players in improving nutrient use efficiency and abiotic stress tolerance
Plant Growth Regulation (2023)
-
Research progress on the biosynthesis and delivery of iron–sulfur clusters in the plastid
Plant Cell Reports (2023)