Abstract
Small G proteins of the Rho family are pivotal regulators of several signaling networks. The Ras homolog family (Rho) and one of its targets, Rho-associated protein kinase (ROCK), participate in a wide variety of biological processes, including bone formation. A previous study has demonstrated that the ROCK inhibitor Y-27632 enhanced bone formation induced by recombinant human bone morphogenetic protein-2 (BMP-2) in vivo and in vitro. However, the effect of other Rho family members, such as Ras-related C3 botulinum toxin substrate 1 (Rac1) and cell division cycle 42 (Cdc42), on bone formation remains unknown. In this study, we investigated whether Rac1 also participates in BMP-2-induced osteogenesis. Expression of a dominant-negative mutant of Rac1 enhanced BMP-2-induced osteoblastic differentiation in C2C12 cells, whereas a constitutively active mutant of Rac1 attenuated that effect. Knockdown of T-lymphoma invasion and metastasis 1 (Tiam1), a Rac-specific guanine nucleotide exchange factor, enhanced BMP-2-induced alkaline phosphatase activity. Further, we demonstrated that BMP-2 stimulated Rac1 activity. These results indicate that the activation of Rac1 attenuates osteoblastic differentiation in C2C12 cells.
Similar content being viewed by others
Main
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor-β (TGF-β) superfamily and participate in various processes associated with the differentiation, growth, and death of cells. BMPs were originally identified as inducers of ectopic bone formation, and they are known to have an important role in bone formation and repair.1, 2 BMPs mediate their effects by binding to type I and II serine/threonine kinase receptors, leading to the activation of the intracellular Smad pathway. The binding of BMPs to their receptors induces the phosphorylation of Smads. Phosphorylated Smads then translocate from the cytoplasm to the nucleus to regulate the transcription of various target genes such as alkaline phosphatase (ALP), runt-related transcription factor 2 (Runx2), and osteocalcin.3
It has been shown that the function of BMPs is modulated by the small GTPase Ras homolog family A (RhoA) and Rho-associated protein kinase (Rho kinase, ROCK).4, 5 We have shown that the expression of a dominant-negative ROCK mutant in mouse stromal ST2 cells induced osteoblastic differentiation, whereas the expression of a constitutively active ROCK mutant attenuated osteoblastic differentiation.4 Continuous supply of a specific ROCK inhibitor (Y-27632) enhanced ectopic bone formation induced by BMP-2.4. Propagating these effects, neogenin acts as a receptor for BMPs and activates RhoA.5 Knockdown of neogenin in mouse C2C12 myoblasts promoted BMP-2-induced osteoblastic differentiation, whereas overexpression of neogenin suppressed this process.5 These findings suggest that RhoA–ROCK signaling negatively regulates BMP-induced osteoblastic differentiation.
The Rho family proteins were identified as Ras-like small GTP-binding proteins. Members of the Rho family, including Rho, Rac, and Cdc42, control the assembly and organization of the actin cytoskeleton in mammalian cells.6 They mediate diverse biological processes, including neuronal morphogenesis, tumor invasion, and bone formation, and act in a coordinated manner to modulate cellular functions.7, 8 However, the effect of Rho family members other than RhoA on osteoblastic differentiation in C2C12 myoblasts remains unclear. In the present study, we assessed the role of Rac1 in BMP-2-induced osteogenesis.
Results
Inhibition of Rac1 promotes BMP-2-dependent osteoblastic differentiation in C2C12 cells
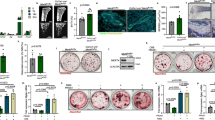
We first examined whether Rac1 activated BMP-2-induced osteoblastic differentiation in C2C12 cells. C2C12 cells were transfected with a dominant-negative mutant of Rac1 (RacDN), a constitutively active mutant of Rac1 (RacCA), or with a control plasmid (mock), with or without rhBMP-2. Thereafter, the cells were stained for ALP (Figure 1a), and the ALP activity was quantified (Figures 1b and c). The BMP-2-induced ALP activity was increased in RacDN-transfected cells. Therefore, inhibition of Rac1 promotes BMP-2-induced osteoblastic differentiation in C2C12 cells. The BMP-2-induced ALP activity tended to decrease in RacCA-transfected cells compared with control cells, although we could not find a significant difference (P>0.05).
Inhibition of Rac1 promotes BMP-2-induced osteoblastic differentiation. (a–c) ALP activity. C2C12 cells were transfected with a dominant-negative form of Rac1 (RacDN), a constitutively active form of Rac1 (RacCA), or with a control plasmid (mock). (a) The cells were cultured with or without rhBMP-2 (300 ng/ml) for 2 days and then stained for ALP. (b, c) Transfected cells were cultured with or without rhBMP-2 (300 ng/ml) for 3 days and then subjected to ALP activity assay. The graphs show fold increase as compared with the levels in the non-treated mock-transfected control group (n=4). (d) Inhibition of Rac1 promotes BMP-2-induced osteoblast gene expression. C2C12 cells were transfected and treated as in (a). mRNA expression of Runx2 (left graph) and osteocalcin (right graph), osteoblastic differentiation markers, was analyzed using real-time PCR. Quantitated mRNA values were normalized to the amount of GAPDH mRNA (n=5). (e) The effect of Cdc42 siRNA on the expression of Cdc42 in C2C12 cells. Cells were transfected with Cdc42 siRNA or control siRNA for 48 h. Western blots for Cdc42 and α-tubulin are shown (upper 2 panels). The signal intensity was quantified by densitometry and normalized to α-tubulin levels (graph: n=3). (f) C2C12 cells were transfected with Cdc42 siRNA or control siRNA. The cells were cultured with or without rhBMP-2 (300 ng/ml) for 4 days and then stained for ALP. For (b–d): *P<0.05, Tukey–Kramer multiple comparison tests. For (e): *P<0.05, unpaired Student’s t test
We then examined the expression of two typical osteogenic marker genes—runt-related transcription factor 2 (Runx2) and osteocalcin—using real-time PCR. BMP-2 treatment enhanced the expression of Runx2 and osteocalcin, and this increase was significantly higher in cells transfected with RacDN than in mock-transfected plasmids (Figure 1d). These results suggest that Rac1 suppressed BMP-2-induced osteoblastic differentiation in C2C12 cells.
Next, we examined the role of Cdc42, another member of the Rho family of proteins, in BMP-2-induced osteoblastic differentiation. C2C12 cells were transfected with short interfering RNA (siRNA) targeting Cdc42. At 48 h post transfection, we confirmed the knockdown of endogenous Cdc42 in C2C12 cells (Figure 1e). After transfection with Cdc42 siRNA, the cells were treated with or without rhBMP-2, and LP activity was examined. The BMP-2-induced ALP activity was increased in Cdc42 siRNA-transfected cells (Figure 1f). Thus, both Cdc42 and Rac1 negatively regulate BMP-2-induced osteoblastic differentiation in C2C12 cells.
Rac1-GEF mediates the inhibition of BMP-2-induced osteoblastic differentiation
Rho family GTPases are activated through interaction with guanine nucleotide exchange factors (GEFs) that regulate their GTP/GDP exchange. We used the Rac1-specific inhibitor NSC23766, which blocks a subset of Rac1 GEFs, Tiam1, and triple functional domain (Trio).9 To examine the role of Trio or Tiam1 in C2C12 osteoblastic differentiation induced by BMP-2, we examined the BMP-2-induced ALP activity in these cells. The cells were pre-treated with NSC23766 at 0, 10, or 50 μM and then cultured with rhBMP-2 for 2 days before being subjected to ALP staining (Figure 2a) or 3 days before ALP activity assay (Figure 2b). The BMP-2-induced ALP activities were increased in NSC23766-treated cells in a dose-dependent manner.
Rac inhibitor enhanced BMP-2-induced osteoblastic differentiation. (a, b) Rac inhibitor NSC23766 enhanced BMP-2-induced ALP activity. C2C12 cells were pre-treated with NSC23766 (0, 10, and 50 μM) for 12 h and then treated with or without rhBMP-2 (300 ng/ml) for 2 days (a) or for 3 days (b). ALP activity was detected by cytochemical staining (a) or quantified by ALP activity assay (b). The graph shows fold increase as compared with the levels in the non-treated control group (n=5). (c) C2C12 cells were pre-treated with or without NSC23766 (50 μM) for 12 h and then incubated with or without rhBMP-2 (300 ng/ml) for 2 days. mRNA expression of Runx2 and osteocalcin was analyzed as in Figure 1d (left graph: n=3; right graph: n=7). (d) MC3T3-E1 cells were pre-treated with or without NSC23766 (50 μM) and then treated with rhBMP-2 (0, 10, and 50 ng/ml) for 3days. (e) Rat Mesenchymal Stem Cells were pre-treated with or without NSC23766 (50 μ M) and then treated with rhBMP-2 (300 ng/ml) for 11 days. The cells were subjected to ALP activity assay as described in (a) and (b). For b–d: *P<0.05, Tukey–Kramer multiple comparison tests
Moreover, we examined the expression of Runx2 and osteocalcin in C2C12 cells cultured with rhBMP-2 in the absence or presence of NSC23766 (50 μ M) for 2 days (Figure 2c). The expression of these osteogenic marker genes was significantly higher in NSC23766-treated cells in the presence of rhBMP-2. These results suggest that Rac1-GEF, Trio, and Tiam1 can suppress BMP-2-induced osteoblastic differentiation via Rac1 activation.
We also examined whether Rac1 has a role in MC3T3-E1 preosteoblastic cells and primary mesenchymal stem cells derived from rat bone marrow. However, NSC23766 treatment did not promote a BMP-2-dependent increase in ALP activities in MC3T3-E1 cells or mesenchymal stem cells (Figures 2d and e).
Tiam1 regulates BMP-2-induced ALP activity
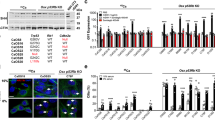
As the Rac1 inhibitor NSC23766 blocks both Tiam1 and Trio, we performed additional experiments to determine which Rac1-GEF was involved in suppressing BMP-2-induced osteoblastic differentiation. In the first set of experiments, C2C12 cells were transfected with siRNA targeting either Tiam1 or Trio. At 48 h post transfection, we confirmed the knockdown of endogenous Tiam1 or Trio in C2C12 cells (Figures 3a and b). BMP-2-induced ALP activity was significantly higher in the cells transfected with Tiam1 siRNA but not in those transfected with Trio siRNA, as assessed by ALP staining (Figures 3c and e) and ALP quantitative assay (Figures 3d and f). These results demonstrated that Tiam1 but not Trio is involved in the suppression of BMP-2-induced ALP activity through Rac1 activation.
Knockdown of Tiam1 enhances BMP-2-induced ALP activity. (a) The effect of Tiam1 siRNA on the expression of Tiam1 in C2C12 cells. Cells were transfected with Tiam1 siRNA or control siRNA for 48 h. Western blots for Tiam1 and α-tubulin are shown (upper two panels). The siRNA did not affect Rac1 expression (lower two panels). The signal intensity was quantified by densitometry and normalized to α-tubulin levels (graph: n=4). (b) The effect of Trio siRNA on the expression of Trio mRNA in C2C12 cells. C2C12 cells transfected with Trio siRNA or control siRNA were cultured for 48 h, and mRNA expression of Trio was examined. The values are normalized to GAPDH (graph: n=3). Trio siRNA did not affect Rac1 expression (lower 2 panels). (c, d) After Tiam1 siRNA transfection, the cells were cultured with or without BMP-2 (300 ng/ml) for 2 days. (c) ALP activity was detected by cytochemical staining. (d) ALP activity was analyzed quantitatively. The graph shows fold increase as compared with the levels in cells treated with control siRNA without rhBMP-2 (n=3). (e, f) After Trio siRNA transfection, the cells were cultured with or without rhBMP-2 (300 ng/ml) for 4 days. (e) ALP activity was detected by cytochemical staining. (f) ALP activity was analyzed quantitatively. The graph shows fold increase as compared with the levels in cells treated with control siRNA without rhBMP-2. For (a, b, d, and f): *P<0.05, Tukey–Kramer multiple comparison tests
BMP-2 induces Rac1 activity in C2C12 cells
Our observation revealed that the inhibition of Rac1 signaling promotes BMP-2-induced osteoblastic differentiation of C2C12 cells.We then examined whether BMP-2 stimulation induced Rac1 activity. We measured the activity of Rac1 by means of a pulldown assay using the GST-fused Rac-binding domain of p21 protein-activated kinase (PAK) beads. Serum-starved C2C12 cells were treated with rhBMP-2 for 1 min. The level of active GTP-bound Rac1 was increased by rhBMP-2 treatment (Figure 4). This result demonstrated that BMP-2 activated Rac1 in C2C12 cells.
BMP-2 stimulates Rac1 activity in C2C12 cells. BMP-2 activates Rac1 in C2C12 cells. Serum-starved C2C12 cells were treated with or without rhBMP-2 (300 ng/ml) for 1 min and subjected to PAK pulldown assays to detect the active form of Rac (upper panels). Whole-cell lysates were immunoblotted with anti-Rac antibody (lower panels). The relative GTP-Rac1 level was normalized to total Rac1 expression and compared with the level in the cells not treated with rhBMP-2 (n=8). For graph: *P<0.05, unpaired Student’s t test
Activation of Rac1 does not inhibit Smad signaling
As the activation of Rac1 negatively regulates BMP-2-induced osteoblastic differentiation in C2C12 cells, we next examined the molecular mechanism underlying the inhibition of BMP signaling by Rac1 activation. BMPs promote bone formation through the activation of Smad signaling.3 Treatment of rhBMP-2 induces phosphorylation of receptor-activated Smads (Smad1, Smad5, and Smad8; Smad 1/5/8). Phosphorylated Smad 1/5/8 translocates into the nucleus to regulate transcription. To investigate whether the activation of Rac1 inhibited Smad signaling, we analyzed the phosphorylation levels of Smad 1/5/8. Serum-starved C2C12 cells were treated with rhBMP-2 in the absence or presence of NSC23766. However, treatment with NSC23766 did not affect the phosphorylation levels of Smad 1/5/8 (Figure 5a). We further examined the effect of Rac1 on Smad localization. C2C12 cells were transfected with RacDN, cultured with or without rhBMP-2, and subjected to immunofluorescence staining to visualize endogenous Smad1 (Figure 5b). The activation of the BMP pathway leads to accumulation of Smad1 in the nucleus (Figure 5b, left lower 2 panels).10, 11 However, cells transfected with RacDN (arrow) were not affected by the localization of Smad1 (Figure 5b, left lowest panel, arrow). These results indicate that other mechanisms might be involved in the inhibition of osteoblastic differentiation through Rac1 activation.
Inhibition of Rac1 does not affect Smad signaling. (a) C2C12 cells were pre-treated with NSC23766 as indicated in Figure 2c and then stimulated with or without rhBMP-2 (300 ng/ml) for 15 min. The phosphorylation levels of Smad 1/5/8 were determined by western blotting (panels). The relative phosphorylation levels of Smad 1/5/8 were normalized by total Smad1 level (graph). The graph showed fold increase as compared with the levels in non-treated cells without rhBMP-2 stimulation. In rhBMP-2-stimulated cells, the relative phosphorylation levels of Smad 1/5/8 in the cells pre-treated with or without NSC23766 were not different. For graph: *P<0.05, Tukey–Kramer multiple comparison tests (n=3). (b) C2C12 cells were transfected with a myc-tagged dominant-negative mutant of Rac1 (RacDN) or with control myc (mock) and treated with or without rhBMP-2 (300 ng/ml). After cell fixation and permeabilization, localization of endogenous Smad1 and myc-tagged proteins was detected using an anti-Smad1 antibody (left panels) and an anti-myc antibody (middle panels). Arrows indicate the cells transfected with myc-tagged RacDN. The nuclei were counterstained with DAPI (right panels). The representative images of cells are shown. Scale bar, 200 μ M. (c) The cells were pre-treated with 3 μ M dorsomorphin and were then treated with rhBMP-2 (300 ng/ml) for 5 min. The phosphorylation levels of Smad 1/5/8 were determined by western blotting. Dorsomorphin inhibited BMP-2-induced phosphorylation of Smad 1/5/8. (d) Dorsomorphin treatment did not affect the expression level of Rac1. (e) Dorsomorphin treatment inhibited the ALP activity induced by Tiam1 siRNA
We further examined whether BMP receptors (BMPRs) were associated with the enhanced ALP activation induced by Rac1 inhibition. C2C12 cells were incubated with a selective inhibitor of BMPR-I, dorsomorphin, for 5 min in the presence or absence of rhBMP-2. rhBMP-2-induced phosphorylation of Smad1/5/8 was inhibited by dorsomorphin (Figure 5c), confirming that dorsomorphin suppresses BMP–Smad signaling. The dorsomorphin treatment did not affect the expression of Rac1 in C2C12 cells (Figure 5d). Enhanced ALP activation induced by Tiam1 siRNA was diminished if the cells were treated with dorsomorphin (Figure 5e). We also demonstrated that the inhibition of Rac1 by itself did not increase the ALP activity in the absence of rhBMP-2 (Figures 1a and 2a). These findings suggest that the BMP–BMPR signal is required for the enhanced ALP activation induced by Rac1 inhibition.
Discussion
In the present study, we assessed the role of Rac1 in osteoblastic differentiation. Inhibition of Rac1 induced the expression of osteoblastic markers such as ALP, Runx2, and osteocalcin. These results indicate that inhibition of Rac1 promotes BMP-2-induced osteoblastic differentiation in C2C12 myoblasts (Figure 6). Although the BMP-2-induced ALP activity was enhanced in RacDN-transfected cells, the ALP activity was not significantly affected by RacCA-transfected cells (Figures 1a–c). The reason why we could not observe significant differences in RacCA-transfected cells could be that the expression of RacCA might not be enough owing to low transfection efficiency. Another possibility is that the activation of Rac1 may only partially inhibit BMP-2-induced osteoblastic differentiation. Rac1 is ubiquitously expressed in most organs and functions to regulate actin cytoskeletal reorganization. Rac1-deficient mice show cell death in numerous locations, particularly in embryonic mesodermal cells.12 Because of embryonic lethality, the role of Rac1 in developmental bone formation remains largely unknown. Our results provide new leads on the effect of Rac1 on osteoblastic differentiation in C2C12 cells.
Rac1, along with RhoA and Cdc42, is one of the most prominent Rho family GTPases. Rho family GTPases regulate actin cytoskeletal reorganization, cell adhesion, and migration. Therefore, they mediate various biological processes such as neuronal morphogenesis, tumor invasion, and bone formation.7, 8 We have previously shown that neogenin, the newly identified receptor for BMP-2, negatively regulates BMP-2 signal transduction through the activation of RhoA.5 BMP-2 binding to neogenin activates RhoA and results in the inhibition of osteoblastic differentiation. Inhibition of RhoA promotes BMP-2-induced osteoblastic differentiation and phosphorylation of Smad 1/5/8. Although RhoA regulates Smad signaling to attenuate bone formation, Rac1 did not affect the phosphorylation levels of Smad 1/5/8 (Figure 5). These observations suggest that Rac1 attenuates BMP-2-induced osteoblastic differentiation through a mechanism that is distinct from RhoA.
Our results show that a dominant-negative mutant of Rac1 promotes osteoblastic differentiation in the presence but not in the absence of BMP-2. Therefore, Rac1 possibly mediates osteoblastic differentiation in a BMP-2-dependent manner. We show that treatment of BMP-2 enhances Rac1 activity in C2C12 cells. Our previous study also demonstrated that BMP-2 induced RhoA activity through neogenin and resulted in suppressed osteoblastic differentiation.5 These findings suggested that BMP-2 not only promotes but also inhibits osteogenesis, although further studies must still be carried out to confirm this. Although it is now possible to generate recombinant human BMPs for medical use, more than 1.5 mg/ml of the recombinant protein is required for bone induction in primates,13 possibly because of their reduced capability for tissue regeneration.
Inhibition of Rac1 did not promote BMP-2-dependent osteoblastic differentiation in the osteoblast-like MC3T3-E1 cells (Figure 2d). Although the mechanism that explains the discrepancy of the results between C2C12 myoblasts and MC3T3-E1 cells is currently unknown and needs further study, the result supports the hypothesis that the role of Rac1 signaling in osteoblastic differentiation varies depending on the cell types or differentiation stages of osteoblasts.
Rac1 is activated through interaction with the diffuse B-cell lymphoma (Dbl) family GEFs, which regulate the exchange of GDP for GTP.14 Dbl family GEFs contain the Dbl-homology domain, which is responsible for GEF catalytic activity, and a pleckstrin-homology domain, which is involved in intracellular targeting. Some of the Rho GEFs show activity toward multiple Rho GTPases, whereas others have more restricted specificity. To determine which GEF is responsible for the activation of Rac1 upon BMP stimulation, we used NSC23766, which can effectively inhibit Rac1-GEF, Trio, and Tiam1. Further, we reveal that siRNA-mediated knockdown of Tiam1 but not Trio induced osteoblastic differentiation. These findings suggest that specific Tiam1/Rac1 interaction participates in BMP-2-induced osteoblastic differentiation in C2C12 cells. Tiam1 is a Rac-specific GEF, whereas Trio contains two GEF domains: one N-terminal domain (TrioN), active on RhoG and Rac1,15 and a C-terminal domain (TrioC), active on RhoA.16 Different specificity to Rho GTPases is a possible explanation of this effect.
In conclusion, we found that inhibition of Rac1 promoted osteoblastic differentiation upon BMP-2 stimulation. Our results provide evidence that Rac1 and Tiam1 are possible molecular targets for promoting osteogenesis.
Materials and Methods
Antibodies and reagents
The following antibodies were used in this study: mouse monoclonal antibodies to Rac (Upstate, Lake Placid, NY, USA) (1:500) and α-tubulin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) (1:1000); rabbit monoclonal antibodies to Smad1 (Cell Signaling Technology, Beverly, MA, USA) (immunocytochemistry; 1:250, western blotting; 1:1000), phospho-Smad1/5/8 (Cell Signaling Technology) (1:1000), Tiam1 (Santa Cruz Biotechnology) (1:1000), and c-Myc (Santa Cruz Biotechnology) (immunocytochemistry; 1:500, western blotting; 1:1000); and Alexa Fluor 568 anti-rabbit IgG and 488 anti-mouse IgG (Life Technologies, Carlsbad, CA, USA). DAPI was purchased from Dojindo Laboratories (Kumamoto, Japan). The recombinant human bone morphogenetic protein-2 (rhBMP-2) was obtained from R&D Systems (Minneapolis, MN, USA). NSC23766, which is a Rac1-specific inhibitor, was obtained from Merck (Darmstadt, Germany). 5′-Adenosine monophosphate-activated protein kinase inhibitor compound C (Dorsomorphin) was purchased from Calbiochem (San Diego, CA, USA).
Cell culture and transfection
C2C12 cells (Riken BRC Cell Bank, Tsukuba, Japan) were cultured in DMEM with 10% FBS. MC3T3-E1 cells (Riken BRC Cell Bank, Tsukuba, Japan) were cultured in α-minimum essential medium (α-MEM) with 10% FBS and 1% penicillin/streptomycin. Rat primary mesenchymal stem cells (Lonza, Walkersville, MD, USA) were cultured using an R-MSCGMBullet Kit (Lonza).
These cells were transfected using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. The cells were assayed 48 or 72 h after transfection. Myc-tagged mouse Rac1-17N and Rac1-61L were kindly gifted by Dr. A. Hall (Department of Biochemistry and Molecular Biology, University College London, London, UK). siRNA against mouse Cdc42, Tiam1, and Trio was obtained from Life Technologies and contained the following sequences:
Cdc42 (5′-GCAAUGAGUGCUAGUUUUUTT-3′)
Tiam1 no. 1 (5′-UUGAGAAUCUGUUCCUCGGUCCUCC-3′),
Tiam1 no. 2 (5′-UUCUGGAGCUGUCUCAGCACUGCUG-3′),
Trio no. 1 (5′-AUCAUUGGCUCGCUUGGGCACGCUG-3′),
Trio no. 2 (5′-UAAAUAGGAAAUGAGCCUCCUGAGG-3′),
Trio no. 3 (5′-UGCACAUGACUUCUACAGCUUUCUC-3′).
The cells were transfected with 10 nM (Cdc42 siRNA, Tiam1 siRNA no. 1), 2.5 nM (Tiam1 siRNA no. 2), or 30 nM (Trio siRNA no. 1, 2, and 3) siRNA using Lipofectamine RNAi Max (Life Technologies) according to the manufacturer’s protocol and cultured for 48 h. Thereafter, the medium was replaced, and the cells were incubated with rhBMP-2 for an additional 48 or 96 h.
Immunocytochemistry
C2C12 cells were fixed with 4% paraformaldehyde for 30 min. The cells were permeabilized, and nonspecific sites were blocked by incubating with blocking solution containing PBS with 0.3% Triton X-100 and 5% BSA. Cells were incubated with the primary antibody diluted in blocking solution for 48 h at 4 °C, washed in PBS, and incubated with Alexa Fluor secondary antibody diluted in blocking solution for 1 h at room temperature. DAPI (1 μg/ml) stain was used to determine nuclear localization. After immunostaining, the slides were mounted with Fluorescent Mounting Medium (DakoCytomation, Glostrup, Denmark).
Assay for Rac activation
After treatment with 300 ng/ml rhBMP-2 or control medium for 1 min, the cells were lysed in a solution containing 25 mM HEPES (pH 7.5), 1% Nonidet P-40, 10 mM MgCl2, 1 mM EDTA, and 1 mM Na3VO4. The cell lysates were clarified by centrifugation (20 400 × g) at 1 °C for 1 min, and the supernatants were incubated with 50 μg of Rac-binding domain of PAK, which had been freshly coupled with glutathione Sepharose-4B beads (GE Health care, Uppsala, Sweden) at 4 °C for 60 min. The beads were washed four times with lysis buffer and subjected to SDS-PAGE, followed by immunoblotting with anti-Rac1 antibody for active Rac1. The cell lysates were also immunoblotted for total Rac1. The levels of Rac1 activation were calculated by comparing the intensities of the active Rac1 bands with those of the total Rac1 bands in each lane using Image J software (National Institutes of Health). The values obtained were then divided by those of the control, and the data are expressed as fold increase in Rac1 activation over control. The construct for the Rac-binding domain of PAK was kindly gifted by Dr. H. Sumimoto (Department of Molecular and Structural Biology, Kyushu University Graduate School of Medical Science, Fukuoka, Japan).
Real-time PCR
Total RNA was extracted from C2C12 cells using the RNeasy kit (Qiagen, Hilden, Germany) and reverse transcribed using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). Approximately 4 μg of total RNA was used as a template to synthesize first-strand cDNA. Subsequently, 1 μl of cDNA mixture was used for real-time PCR (total volume, 20 μl) performed using SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions. In addition, the PCR products were purified and sent for sequence analysis using a 7300 Real-time PCR system (Applied Biosystems) to verify amplification of the targeted genes. The optimal conditions were defined as follows: 40 cycles at 50 °C for 2 min, at 95 °C for 10 min, at 95 °C for 15 s, and at 60 °C for 1 min, followed by 1 cycle at 95 °C for 15 s, at 60 °C for 30 s, at 95 °C for 15 s, and at 60 °C for 15 s. The relative mRNA expression of the markers of osteoblastic differentiation was adjusted according to the expression of GAPDH. The primer pairs used for PCR are as follows: Runx2, forward 5′-GCTTGATGACTCTAAACCTA-3′, reverse 5′-AAAAAGGGCCCAGTTCTGAA-3′; Osteocalcin, forward 5′-CTCACTCTGCTGGCCCTG-3′, reverse 5′-CCGTAGATGCGTTTGTAGGC-3′; Trio forward 5′-GCTGTCACGGCTAGAGGAAC-3′, reverse 5′-CGAGCCTGAGTTCTTTTTGG-3′; and GAPDH, forward 5′-TGAACGGGAAGCTCACTGG-3′, reverse 5′-TCCACCACCCTGTTGCTGTA-3′. The specificity of each primer set was determined with a pre-test showing amplification of a specific gene by gel visualization and sequencing. The results were analyzed and validated using the relative standard curve method and the delta-delta Ct method.
ALP staining
C2C12 cells were plated on 12-well plates and treated with NSC23766 for 12 h or transfected with the indicated expression vectors for 48 h. The culture medium was replaced with differentiation medium, which was DMEM containing 10% FBS, 10 μg/ml ascorbic acid 2-phosphate (Wako, Osaka, Japan), and 5 mM β-glycerophosphate (Merck KGaA, Darmstadt, Germany) with or without rhBMP-2. The cells were cultured for 2 or 3 days (indicated in the figure legends). MC3T3-E1 cells were plated on 12-well plates and treated with NSC23766 for 12 h. The culture medium was α-MEM containing 10% FBS and 1% penicillin/streptomycin with or without rhBMP-2. The MC3T3-E1 cells were cultured for 3 days. Rat Mesenchymal Stem Cells were plated on 12-well plates and treated with NSC23766 for 12 h. The cells were cultured for 11 days using an R-MSCGMBullet Kit with or without rhBMP-2. These cells were fixed for 15 min with 4% formaldehyde/PBS at room temperature. They were washed with PBS and then stained with nitro blue tetrazolium (NBT; Promega, Madison, WI, USA) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP; Promega) in ALP buffer (100 mM of Tris-HCl [pH 9.5], 100 mM NaCl, and 5 mM MgCl2).
ALP activity assay
C2C12 cells were plated on 12-well plates and treated with NSC23766 for 12 h or transfected with the indicated expression vectors or siRNA for 48 h. The culture medium was replaced with the differentiation medium described above. Cells were cultured for 2 or 3 days (indicated in the figure legends) and then lysed in Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. ALP activity was assayed using p-nitrophenylphosphate as a substrate in the LabAssay ALP (Wako), and the protein content was measured using the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific).
Western blotting analysis
The siRNA-transfected or rhBMP-2-stimulated C2C12 cells were washed in PBS. After washing the cells, cell lysis was carried out using a lysis buffer containing 50 mM Tris (pH 7.4), 1% Noniet P-40, 150 mM NaCl, and complete Protease Inhibitors (Roche Applied Science, Mannheim, Germany). When we analyzed the phosphorylation levels of Smad 1/5/8, 10 mM NaF and 10 mM Na3VO4 were added to the lysis buffer. The proteins were separated by SDS-PAGE followed by immunoblotting.
Statistical analysis
The quantitative data are expressed as mean±SEM of at least three (indicated in the figure legends when the number was more than three) independent experiments. Statistical analysis of these values was performed using one-way analysis of variance followed by Tukey–Kramer multiple comparison tests or an unpaired Student’s t test (Figures 1e and 4). P values of less than 0.05 were considered significant.
Abbreviations
- Rho:
-
Ras homolog family; Rho kinase
- ROCK:
-
Rho-associated protein kinase
- BMP:
-
bone morphogenetic protein
- Rac1:
-
Ras-related C3 botulinum toxin substrate 1
- Cdc42:
-
cell division cycle 42
- Tiam1:
-
T-lymphoma invasion and metastasis 1
- TGF-β:
-
transforming growth factor-β
- ALP:
-
alkaline phosphatase
- Runx2:
-
runt-related transcription factor 2
- RhoA:
-
Ras homolog family A
- GEFs:
-
guanine nucleotide exchange factors
- Trio:
-
triple functional domain
- PAK:
-
p21 protein-activated kinase
- JNK:
-
c-Jun N-terminal kinase
References
Urist MR . Bone: formation by autoinduction. Science 1965; 150: 893–899.
Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW et al. Novel regulators of bone formation: molecular clones and activities. Science 1988; 242: 1528–1534.
Miyazono K . A new partner for inhibitory Smads. Cytokine Growth Factor Rev 2002; 13: 7–9.
Yoshikawa H, Yoshioka K, Nakase T, Itoh K . Stimulation of ectopic bone formation in response to BMP-2 by Rho kinase inhibitor: a pilot study. Clin Orthop Relat Res 2009; 467: 3087–3095.
Hagihara M, Endo M, Hata K, Higuchi C, Takaoka K, Yoshikawa H et al. Neogenin, a receptor for bone morphogenetic proteins. J Biol Chem 2011; 286: 5157–5165.
Hall A . G proteins and small GTPases: distant relatives keep in touch. Science 1998; 280: 2074–2075.
Etienne-Manneville S, Hall A . Rho GTPases in cell biology. Nature 2002; 420: 629–635.
Takai Y, Sasaki T, Matozaki T . Small GTP-binding proteins. Physiol Rev 2001; 81: 153–208.
Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y . Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA 2004; 101: 7618–7623.
Lagna G, Hata A, Hemmati-Brivanlou A, Massague J . Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature 1996; 383: 832–836.
Liu F, Hata A, Baker JC, Doody J, Carcamo J, Harland RM et al. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature 1996; 381: 620–623.
Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H et al. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene 1998; 17: 3427–3433.
Govender S, Csimma C, Genant HK, Valentin-Opran A, Amit Y, Arbel R et al. Recombinant human bone morphogenetic protein-2 for treatment of open tibial fractures: a prospective, controlled, randomized study of four hundred and fifty patients. J Bone Joint Surg Am 2002; 84: 2123–2134.
Zheng Y . Dbl family guanine nucleotide exchange factors. Trends Biochem Sci 2001; 26: 724–732.
Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouviere C, Fort P . TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of rhoG. J Cell Sci 2000; 113: 729–739.
Debant A, Serra-Pages C, Seipel K, O'Brien S, Tang M, Park SH et al. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc Natl Acad Sci USA 1996; 93: 5466–5471.
Acknowledgements
We thank Dr. A. Hall for providing us plasmids expressing Rac1-17 N and Rac1-61L. We are also grateful to Dr. H. Sumimoto for plasmid expressing the Rac-binding domain of PAK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by Y Shi
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Onishi, M., Fujita, Y., Yoshikawa, H. et al. Inhibition of Rac1 promotes BMP-2-induced osteoblastic differentiation . Cell Death Dis 4, e698 (2013). https://doi.org/10.1038/cddis.2013.226
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.226
Keywords
This article is cited by
-
Prognostic significance of long non-coding RNA five prime to XIST in various cancers
BMC Cancer (2022)
-
MiR-1224-5p modulates osteogenesis by coordinating osteoblast/osteoclast differentiation via the Rap1 signaling target ADCY2
Experimental & Molecular Medicine (2022)
-
MiR-93 inhibits the vascular calcification of chronic renal failure by suppression of Wnt/β-catenin pathway
International Urology and Nephrology (2022)
-
Adaptor protein CrkII negatively regulates osteoblast differentiation and function through JNK phosphorylation
Experimental & Molecular Medicine (2019)
-
Mechanically-sensitive miRNAs bias human mesenchymal stem cell fate via mTOR signalling
Nature Communications (2018)