Abstract
Ras proteins undergo an incompletely understood trafficking process in the cell. Rasosomes are protein nanoparticles of 80–100 nm diameter that carry lipidated Ras isoforms (H-Ras and N-Ras) as well as their effectors through the cytoplasm and near the plasma membrane (PM). In this study, we identified the subcellular origin of rasosomes and how they spread Ras proteins through the cell. We found no dependency of rasosome formation on galectins, or on the GDP-/GTP-bound state of Ras. We found that significantly more rasosomes are associated with forms of Ras that are localized to the Golgi, namely N-Ras or the singly palmitoylated H-Ras mutant (C181S). To explore the possibility that rasosome originate from the Golgi, we used photoactivatable (PA)-GFP-H-Ras mutants and showed that rasosomes bud from the Golgi in a two-step mechanism. Newly released rasosomes first move in an energy-dependent directed fashion and then convert to randomly diffusing rasosomes. Dual fluorescence time-lapse imaging revealed the appearance of dually labeled rasosomes, indicating a dynamic exchange of cytoplasmic and PM-associated Ras with rasosome-associated Ras. Finally, higher levels of rasosomes correlate with higher levels of ERK phosphorylation, a key marker of Ras downstream signaling. We suggest that H-Ras and N-Ras proteins exchange with rasosomes that can function as carriers of palmitoylated Ras and its signals.
Similar content being viewed by others
Main
Ras proteins are small GTPases that cycle between an inactive GDP-bound state and an active GTP-bound state to control cell proliferation, differentiation, survival and death.1, 2, 3, 4, 5 Active GTP-bound Ras triggers multiple downstream effectors that lead to activation of diverse signaling cascades resulting in changes in cellular behavior.4, 6 Ras proteins are mutationally activated in as many as 30% of all human tumors,5, 7, 8 and Ras is an important target for cancer therapy.
Since Ras activation occurs predominantly at the plasma membrane (PM), it was generally considered that Ras signals originate exclusively from this region. However, recently it was shown that stable pools of endomembrane-associated Ras (i.e., Golgi- and ER-associated Ras), also serve as Ras signaling platforms.9, 10 A major question about compartmentalized Ras signaling is how activating signals from the PM are relayed to endomembranes, through the cytoplasm. All Ras isoforms (H-, K-, and N-Ras) undergo an irreversible farnesylation of a cysteine residue.11 H-Ras and N-Ras require an additional lipid modification, palmitoylation, for their stable anchoring to the PM: H-Ras undergoes double palmitoylation (at cysteines 181 and 184), whereas N-Ras undergoes single palmitoylation (at cysteine 181).12, 13, 14 These modifications are important for Ras membrane targeting, Ras signaling regulation, and intracellular trafficking.15 The palmitoylation-dependent trafficking pathway, also termed the acylation cycle,16 is a cycle of palmitoylation and depalmitoylation reactions that determines a constant transport of H-Ras and N-Ras between the Golgi and the PM. Palmitoylation at the Golgi promotes the association of Ras with the PM, whereas depalmitoylation releases the protein from the PM.16
It is still unclear exactly how palmitoylated Ras (highly hydrophobic) trafficks from the Golgi through the hydrophilic cytoplasm, or how depalmitoylated (but farnesylated) Ras travels from the PM to the Golgi. Possible modes of trafficking include both vesicular15, 16 and nonvesicular15, 16, 17 mechanisms. Recently, it was suggested that cGMP phosphodiesterase 6δ (PDE6δ) acts as a chaperone for all Ras isoforms, and can serve as a GDI-like solubilizing factor for hydrophobic Ras in the cytoplasm by binding the farnesyl moiety, although palmitoylated H-Ras binds poorly to it.18 At the PM, Ras is escorted by other proteins, essential for its proper anchoring: galectin-1 and galectin-3, escort H-Ras, and K-Ras, respectively,19, 20, 21 whereas N-Ras is escorted by nucleolin.22
Another mode of intracellular Ras trafficking involves rasosomes;23 rapidly diffusing nanoparticles of about 80–100 nm in diameter, that carry Ras proteins and their effectors (e.g., ERK and p-ERK).23 In order to be loaded on rasosomes, Ras must possess at least one palmitoyl moiety;23 therefore, H-Ras and N-Ras but not K-Ras(4B) can be carried by rasosomes. Rasosomes do not overlap with exocytic pathway vesicles and their motion is independent of ATP and the cytoskeleton.23 Further, the Ras-binding domain of the Ras effector Raf-1 is recruited to rasosomes upon EGF stimulation, suggesting that either Ras is activated on the rasosome or that active Ras can be loaded onto it.24 However, the origin of rasosomes is not known. Here, we determine the cellular origin of rasosomes, show how rasosomes can spread Ras across the cell and show association between rasosomes and ERK activation.
Results
Rasosome formation is independent of the escort proteins Galectin-1 and -3
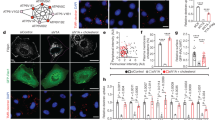
Galectin-1 (Gal-1) is a critical escort protein for H-Ras, whereas Galectin-3 (Gal-3) primarily escorts K-Ras.19, 20 Interaction with Gal-1 is crucial for H-Ras localization and signaling from PM nanoclusters.25, 26 To determine whether there is an association or dependency between galectins and rasosomes, we expressed GFP-H-Ras in mouse embryonic fibroblast (MEF) cell lines derived from knockout mice lacking Gal-1, Gal-3, or both (double knockout, DKO).27 We imaged and recorded the location of GFP-Ras in live cells. The vast majority of cells displayed rasosomes regardless of their genotype (Figure 1a). These results indicate that Galectin-1 and Galectin-3, which are escort proteins for H-Ras and K-Ras, respectively, and support Ras nanocluster formation and maintenance in the PM,28 are not required for rasosome formation.
Number of rasosomes is enhanced in Ras isoforms that have preferential Golgi localization compared with Ras isoforms that have preferential PM localization. (a) Four MEF cell lines (WT, Gal 1 −/−, Gal 3 −/−, and Gal 1/3 −/−) expressing GFP-H-Ras-WT were imaged by epifluorescence (epi) microscopy. The percentage of cells expressing Rasosomes was calculated in two independent experiments (86.3±5.3% in WT MEFs, 88.8±2.8% in Gal-1−/− MEFs, 80.5±5.5% in Gal-3−/− MEFs, and 85.5±5.5% in DKO MEFs, n≥ 12 in each). No significant difference in the percentage of cell expressing rasosomes between all four cells lines was found (AVOVA). (b) COS-7 cells expressing GFP-H-Ras-WT (n=30), GFP-H-Ras-12V (n=31), or GFP-H-Ras- S17N (dominant negative) (n=27) were imaged by epifluorescence (epi) microscopy. The amount of rasosomes was measured and calculated using the MF algorithm, and no statistical differences were found between GFP-H-Ras-WT, GFP-H-Ras-12V, and GFP-H-Ras- S17N (left panel, P=0.191, Kruskal–Wallis test). Similarly, no statistical differences in the amount of rasosomes was found between GFP-N-Ras-WT (n=22) and GFP-N-Ras-G13V (n=23) (right panel; P=0.265, t-test). (c) Cells expressing GFP-H-Ras-WT (n=14) or GFP-N-Ras-WT (n=13) were imaged by TIRF microscopy and the amount of rasosomes was determined as in (a). Cells expressing GFP-N-Ras-WT had significantly more rasosomes than cells expressing GFP-H-Ras-WT (*P=0.033, t-test). (d) Representative images of a cell expressing GFP-H-Ras-WT (left image, low levels of rasosomes), a cell expressing GFP-H-Ras-C181S (middle image, high levels of rasosomes and a cell expressing GFP-H-Ras-C184S (right image, low levels of rasosomes). Single rasosomes are indicated with arrows. (e) Statistical analysis of experiments performed as described in (a) with cells expressing GFP-H-Ras-WT (n=34), GFP-H-Ras-C181S (n=34), and GFP-H-Ras-C184S (n=32). C181S-expressing cells showed significantly higher amounts of rasosomes than the other cells (P<0.0001, Kruskal–Wallis test) (left panel). Similar results were obtained at the TIRF plane (***P<0.0001, Kruskal–Wallis test) (right panel). (f) Mean GFP fluorescence intensities in the same regions where rasosomes were measured in (d) by epi and TIRF microscopy (all three Ras variants). No significant differences were found in the cytoplasm (epi) (P=0.2753, one-way ANOVA); however, GFP-H-Ras-C184S showed higher fluorescence compared with all other constructs at the TIRF plane (**P=0.0024, one-way ANOVA). Western blot analysis shows that the levels of expression were similar between GFP-H-Ras-WT and GFP-H-Ras-C181S (P=0.46, t-test). Bars represent mean±S.E.
Rasosome formation does not depend on the GDP-/GTP-bound state of Ras
Given that Ras interacts with different nanodomains on the PM depending on its activation state,23, 29, 30, 31, 32, 33 we examined whether rasosome formation is influenced by the GDP-/GTP-bound state of Ras. For this purpose, we transfected cells with GFP-tagged Ras locked in different activation states. First, we performed time-lapse imaging on cells expressing H-Ras-WT (that can cycle between GDP- and GTP-bound states and associates dynamically with lipid rafts), H-Ras-12V (a chronically GTP-bound and active mutant that is excluded from lipid rafts but associated with saturable non-raft sites) or H-Ras-S17N (a chronically GDP-bound and inactive dominant negative (DN)) (Figure 1b). To quantitate the amount of rasosomes observed under each condition, we then analyzed the movies using our mobility factor (MF) algorithm24 (see Materials and methods). This algorithm gives a numerical representation of the extent of mobile particles per area, reflecting the amount of rasosomes.24 We found that the GDP-/GTP-bound state of H-Ras does not influence the number of rasosomes formed, compared with WT Ras (Figure 1b, left panel). In a similar experiment performed on cells expressing N-Ras-WT or N-Ras-G13V (a chronically GTP-bound and active mutant, preferentially associated with rafts31) (Figure 1b, right panel), we also found no significant difference in rasosome formation. Hence, the expression of different Ras mutants that target H-Ras and N-Ras to either raft or non-raft domains at the PM caused no change in the number of the rasosomes; suggesting that the microlocalization of Ras within the PM, which is determined by the GDP-/GTP-bound state of Ras34, 35, 36 has no effect on rasosome formation.
More rasosomes are present in N-Ras than in H-Ras expressing cells
While H-Ras is targeted mainly to the PM with some Golgi localization, N-Ras is scattered equally between the PM and the Golgi.35 We examined whether there is a preferential loading of N-Ras or H-Ras onto rasosomes. Therefore, we directly compared the rasosomes formed upon expression of GFP-tagged N-Ras or H-Ras. Based on our MF analysis, cells expressing N-Ras-WT showed a significantly higher number of rasosomes compared with the cells expressing H-Ras-WT (P=0.033) (Figure 1c). Thus, the singly palmitoylated N-Ras that is more Golgi-oriented is associated with more rasosomes than the dually palmitoylated and less Golgi-oriented H-Ras. This suggests that either an extra palmitoyl moiety reduces Ras association with rasosomes, or that higher levels of Golgi-associated Ras result in higher levels of rasosomes, or both.
A singly palmitoylated and Golgi-targeted mutant of H-Ras generates more rasosomes than H-Ras-WT
To distinguish the contributions of the number of palmitate moieties versus Golgi localization, we generated two different mutants of GFP-H-Ras, each containing a single palmitoylation site (H-Ras-C181S, designated C181S; and H-Ras-C184S, designated C184S). It is known that the C181S mutant is localized mostly to the Golgi whereas the C184S mutant is localized mostly to the PM.35 Imaging of the cytoplasm and PM regions with epifluorescence and TIRF microscopy, respectively, visually demonstrated the presence of more rasosomes in the cells expressing C181S compared with H-Ras-WT and C184S (representative images are shown in Figure 1d). We then quantified rasosomes amount and found that cells expressing C181S exhibited significantly higher numbers of rasosomes (Figure 1e), both near the PM (∼7.2-fold higher) and in the cytoplasm (∼6.5-fold higher) compared with cells expressing either H-Ras-WT or C184S. We also used GTP-bound versions of Ras and found that cells expressing GFP-H-Ras-12V-C181S exhibited significantly more rasosomes than the others (P<0.0001, Kruskal–Wallis test, for both epifluorescence and TIRF recordings, data not shown).
We ruled out the trivial explanation that the higher number of rasosomes (in C181S-expressing cells) might be due to higher expression levels compared with other H-Ras proteins, both by performing western blot and by quantification of the overall GFP fluorescence (Figure 1f).
As indicated above, targeting of C181S to the PM is low compared with that of C184S. Nevertheless, we observed significantly more rasosomes at the PM of cells expressing C181S compared with cells expressing C184S (P<0.0001, Figure 1e, TIRF plane). Collectively, these experiments suggest that rasosome formation correlates with the amount of Ras in the Golgi and not in the PM.
Rasosomes bud from the Golgi
Because we observed a strong correlation between the levels of rasosomes and Golgi localization of Ras, we next examined the possibility that rasosomes originate from the Golgi. To do this, we imaged cells expressing photoactivatable (PA) GFP-H-Ras-C181S. To locate the Golgi, cells were co-transfected with mRFP-H-Ras-WT. Once the Golgi region was identified, we photoactivated the PA-GFP-H-Ras-C181S specifically at the Golgi. No rasosomes or other organelles were detected near the Golgi area immediately following activation (Figure 2a). To minimize bleaching we continued to image the cells at 1 Hz for about 1.5 min, and then we imaged at 9 Hz for an additional 5 s. We observed fluorescent particles budding from the Golgi (Figure 2a), tracked single particles, and found that they initially move in a directed motion that later turns into a rapid Brownian motion throughout the cell (Figure 2b). We analyzed23 the motion of single particles after they began to move randomly and found that these particles exhibit the same motion characteristics as rasosomes. The calculated diffusion coefficients of these particles were at the order of magnitude of 10−2 μm2/s, in agreement with diffusion coefficients of GFP-H-Ras rasosomes described previously (D ranges between 10−2–10−1 μm2/s23).
Rasosomes bud from the Golgi. COS-7 cells co-transfected with mRFP-H-Ras-WT and PA-GFP-H-Ras-C181S were imaged about 24 h after transfection. The Golgi region was detected using the mRFP, and the PA-GFP-H-Ras-C181S was photoactivated in this region (appears as the big fluorescent area in (a)). (a) A typical cell before and after PA-GFP activation. Time past activation (in seconds) is indicated. The first six images (upper panel) were taken at a rate of 1 Hz and represent the early particle’s directed motion. The last three images (lower panel) were taken at 9 Hz and represent the subsequent random diffusion part of the particle’s motion. Red circle marks the same single rasosome. (b) Trajectories of the particle shown in (a). On the left, trajectory from the early images representing the directed motion of the rasosome (time scale is 80 s). Note that at the end of the movie (red color) the particle starts to move randomly. On the right, trajectory from the fast acquired movie, representing the subsequent particle’s random motion (time scale is 2 s). The trajectories are color coded by time where the blue is the starting point and red in the ending point. The black circle represent the starting point, its diameter is 200 nm
Previously, we showed that the movement of rasosomes in the cytoplasm is temperature-independent.23 We repeated the photoactivation experiments at both normal (35 °C) and low (17 °C) temperatures to examine temperature effect on rasosomes budding. Photoactivation at 35 °C, within the Golgi region of cells expressing PA-GFP-H-Ras-C181S, results in rasosome spreading from the Golgi towards the cell periphery, as shown above. At 17 °C, although photoactivation lights up the Golgi region, the budding of rasosomes is severely compromised (3.555±0.8 rasosomes bud over 2 min at 35 °C; 0.19±0.1 rasosomes bud at 17 °C; P<0.001, t-test). Hence, the initial step of rasosome budding from the Golgi is temperature-dependent, unlike the later randomized movement in the cytoplasm. This temperature dependence suggests that rasosome budding involves some motor activity that allows the initial directed motion and is inhibited at low temperature, similar to vesicular trafficking.
Rasosome-associated Ras undergoes dynamic exchange with cytoplasmic and PM Ras
To test whether Ras is permanently or temporarily loaded on the rasosomes, we utilized PA-GFP-tagged Ras. Because we observed that rasosomes bud from the Golgi, we performed these experiments with PA-GFP-N-Ras that is most prominently present there. We applied two excitation paradigms to differentiate between exchange and no exchange. First, we excited only a small area, about 20% of the cell (‘small area’). We expected that, if there is an exchange of Ras between the small amount of the rasosomal photoactivated Ras and the large amount of non-photoactivated cytoplasmic Ras, the intensity and number of fluorescently labeled rasosomes will be reduced over time (see scheme in Figure 3a). Alternatively, we excited the entire cell, thereby photoactivated most of the Ras proteins in it (‘whole cell’). In this case, any exchange of rasosome-associated Ras would not produce decreased fluorescence intensity or fewer fluorescent rasosomes. Instead, signal decay would more likely be due to photobleaching (serves as control).
Rasosome-associated Ras undergoes dynamic exchange with cytoplasmic Ras. (a) A schematic representation of the experimental logic: PA-GFP-N-Ras-WT was photoactivated in the whole cell (left panel) or in a small spot in the cell (right panel). In the partial activation, one of the two possibilities is expected: (i) if rasosomal Ras exchanges with other Ras proteins in the cell, we will observe a decrease in the GFP fluorescence and amount of the rasosomes (left arrow), since the cytoplasmic Ras is mostly ‘dark’ (non-activated PA-GFP), (ii) if rasosomal Ras does not exchange with its surroundings, we will observe no change in rasosomes fluorescence (right arrow). When the whole cell is activated (serving as control) we do not expect any change. (b) Images of a partly activated cell; blue line marks cell’s boundaries, yellow circle marks the photoactivated area. (c) The calculated value of rasosomes (MF) at t=5 and t=15 min for cells that were partly activated (left panel, n=14, t-test, P=0.028) were reduced over time. No significant decrease in MF was observed in fully activated cells (right panel, n=18, P=0.27, t-test); bars represent mean values±S.E. (d) We co-transfected cells with mRFP-H-Ras-WT and PA-GFP-H-Ras-WT (t=0) and simultaneously activated the GFP and bleached the mRFP at a small area (t=1 s). Then, we imaged the PM with TIRF microscopy and observed GFP-Ras being loaded on the mRFP-rasosomes (white arrow, t=3 s).
We calculated the MF index at different times after photoactivation, and compared the values for the ‘small area’ and the ‘whole cell’. The MF decreased between t=5 min and t=15 min in the ‘small area’ photoactivation (Figures 3b and c, left panel) but not in the ‘whole cell’ photoactivation (Figure 3c, right panel). Therefore, we suggest exchange between photoactivated Ras and the non-fluorescent Ras in its surroundings.
Additionally, we examined whether exchange of Ras and rasosomes occurs near the PM. For these experiments we used two versions of H-Ras (mRFP-H-Ras and PA-GFP-H-Ras), which is more prominently localized to the PM than N-Ras. We simultaneously photobleached the red Ras and activated the green Ras on the same rasosome. Upon recovery, if exchange of Ras between rasosome and the surrounding Ras occurs, we expect red rasosomes outside the bleached area acquiring green Ras, and green rasosomes in the photoactivated area acquiring red Ras. We imaged the cells at the TIRF plane and first observed red-labeled mRFP-H-Ras rasosomes near the cell membrane, with no GFP fluorescence (Figure 3d, 0 s). After photobleaching and photoactivation we observed the appearance of a spot of green fluorescent Ras and rasosomes and a complete disappearance of the red mRFP-H-Ras and red-labeled rasosomes within this spot (Figure 3d, 1 s). Hence, no two-colored population of rasosomes was detected at this time. However, within a few seconds, mRFP rasosomes were being loaded with GFP, creating two-colored rasosomes (Figure 3d, t=3 s). These results suggest that H-Ras undergoes exchange on rasosomes near the PM.
High levels of ERK phosphorylation correlate with high levels of rasosomes
To determine whether rasosomes exhibit a biological function, we compared the phosphorylation levels of two key Ras downstream effectors in cells expressing H-Ras-WT and C181S. Although the proteins differ by only one amino acid (C181S), they significantly differ in the level of rasosomes (Figure 1e). We compared ERK and AKT phosphorylation in lysates of cells expressing either GFP-H-Ras-WT or C181S. We found a significant increase (46±4.5%) in the amount of p-ERK in the C181S cells (normalized to the levels of total ERK in the cells) compared with the WT cells (n=5, P<0.05, t-test) (Figure 4a). However, we did not detect any difference in AKT phosphorylation (Figure 4b). The positive correlation seen here between rasosomes and p-ERK is consistent with the possibility that rasosomes serve as signaling platforms for the Ras-MAPK pathway.
ERK phosphorylation is increased in C181S cells. Western blot experiments were performed with triplets of lysates from COS-7 cells that were transfected either with GFP-H-Ras-C181S or with GFP-H-Ras-WT. (a) To compare the levels of ERK phosphorylation between cell types we calculated the ratio of p-ERK/ERK. C181S-expressing cells show higher levels of ERK phosphorylation (*P<0.05, t-test). Left panel: representative blots. Right panel: densitometry. (b) Similar measurements found no significant differences in the ratio of p-AKT/AKT (t-test). Left panel: representative blots. Right panel: densitometry. Bars represent mean values±S.E.
Discussion
Rasosomes serve as carriers of the hydrophobic H- and N-Ras across the cytoplasm
Our results strongly suggest that rasosomes bud from the Golgi and move towards the PM. Previous studies showed that Ras depalmitoylation occurs in the PM and repalmitoylation occurs in the Golgi.16 Thus, rasosomal Ras is highly hydrophobic as it is modified irreversibly by a farnesyl group and also modified reversibly by at least one palmitoyl moiety. Thus, none of the proposed models can provide a definitive answer to the question of how hydrophobic Ras traffics rapidly through the hydrophilic cytoplasm. Recently, it has been suggested that PDE6δ carries farnesylated H-Ras and N-Ras through the cytoplasm.18 PDE6δ binds preferentially to depalmitoylated Ras and was proposed to act as a solubilization factor of H-Ras, N-Ras, and K-Ras. The trafficking of palmitoylated Ras from the Golgi to the PM may occur by vesicles15, 16, 37, 38, 39 although nonvesicular traffic has also been demonstrated.15, 30 Our results show an additional mode that allows transfer of palmitoylated H-Ras and N-Ras from the Golgi to the PM via rasosomes. Also we show that the H-Ras mutant C181S creates the highest amount of rasosomes relative to the other palmitoylated Ras proteins, this complements the recent results which shows that this mutant has the lowest tendency to be solubilized by PDE6δ.18 Hence, rasosomes can act as an alternative mode for transfer of hydrophobic Ras that cannot be carried by PDE6δ.
Rasosomes association with the PM and with the Golgi
The anchoring of H-Ras to the PM and its association with nanoclusters on the PM is supported by galectins28 and is affected by the GDP/GTP state of Ras. We found that lack of galectins had no effect on the number of rasosomes, and that both H-Ras and N-Ras association with rasosomes is independent from their GDP-/GTP-bound state that determines the microlocalization of Ras in the PM. Interestingly, escorting of Ras by PDE6δ, is also not affected by the GDP-/GTP-bound state of Ras. The lack of depandency on galectins or the GDP/GTP-binding state reduces the likelihood that formation of rasosomes is dependent on Ras PM nanoclusters, which themselves depend on galectins, and thus our findings that rasosome formation correlates with localization of Ras in the Golgi complement these findings and strengthen our conclusion that rasosomes originate from the Golgi. This also supports a possible connection between rasosomes and the acylation cycle in which both H-Ras and N-Ras cycle between the Golgi and the PM independent of their GDP-/GTP-bound state.16
Additionally, N-Ras transfected cells exhibit more rasosomes than H-Ras transfected cells. The two main differences between these isoforms are that (i) N-Ras is mostly targeted to the Golgi and H-Ras is localized mostly to the PM35 and that (ii) H-Ras has two palmitoyl modifications while N-Ras has only one. In addition, in the C181S mutation, which has single palmitoyl and is Golgi targeted like N-Ras, more rasosomes are created than in H-Ras-WT or the singly palmytoylated C184S. These findings join previous studies that show that too much lipid glue may even stop Ras signaling.40, 41, 42 In the case of rasosomes, it seems that not only the valence of palmitoylation is important but also its location.
Rasosomes are formed at the Golgi
To further investigate the origin of the rasosomes, we utilized photoactivation of PA-GFP-Ras constructs. We found Ras containing particles that bud from the Golgi in a motion that seems to be first directed and then becomes random, similar to rasosomes. Therefore, we hypothesize that rasosomes are first budding from the Golgi and then travel to the peripheral areas of the cell and to the PM. The vesicular transport of H-Ras from the Golgi towards the PM has been already described.37, 39 Our new findings suggest, however, that rasosomes are an alternative mechanism of Ras transport from Golgi to the PM. This mechanism may also involve retrograde transport from the PM to the Golgi allowing communication between the PM and Golgi Ras and vice versa. Our finding that the budding of rasosomes from the Golgi is inhibited by low temperature, similar to the budding of Golgi vesicles, suggests that the early step of rasosome budding is energy-dependent, while the later random movement is not.
The biological function of Rasosomes
Previous studies have shown that ERK activation induced by H-Ras-12V tethered by a specific Golgi-targeting signal is as strong as the signal activated by normally distributed H-Ras-12V.9 Our results indicate another property of Ras/ERK signaling that appears to be associated with rasosome trafficking. We found that ERK phosphorylation is increased in C181S-expressing cells compared with GFP-H-Ras-WT expressing cells. The main differences between these cells are in Ras localization and in the amount of rasosomes. While H-Ras-WT is localized mainly in the PM and generates a small number of rasosomes, C181S is localized mainly in the Golgi and exhibits a high number of rasosomes. Compared with the earlier studies,9 our experiment demonstrates that Golgi-associated Ras signals much stronger to the ERK cascade than the PM-targeted Ras. Thus, the increase in ERK phosphorylation is likely due to the high levels of rasosomes. Notably, this increase occurs even without EGF stimulation or presence of 12V mutation, therefore, the higher p-ERK observed in these cells is highly correlated with the higher levels of rasosomes.
Interestingly unlike Ras association with rasosomes, the membrane association of Ras proteins is strongly enhanced by escort proteins such as galectins and nucleolin20, 22 and not by PDE6δ. Neither Gal-1 nor Gal-3 are detected in rasosomes (unpublished observations) supporting the notion that these proteins do not escort Ras in the rasosomes. Indeed, Gal-1 KO, Gal-3 KO, and the double KO cells exhibited normal levels of rasosomes.
Rasosomes spread palmitoylated, Golgi-originated Ras across the cell
Our studies show that Ras in rasosomes undergoes a dynamic exchange with other Ras pools in the cell and that both H-Ras and N-Ras participate in this process. Since we found that this dynamic exchange occurs both at the cytoplasm and near the PM, and since rasosomes are budding from the Golgi, we suggest a model by which rasosomes allow the two main cellular Ras pools, that is, Golgi Ras pool and PM Ras pool, to communicate and exchange their Ras. Rasosomes loaded with palmitoylated Ras bud from the Golgi to the cytoplasem towards the PM, where they can exchange their loaded Ras. This is an alternative mechanism that enables Golgi Ras to travel to the PM, where it can undergo activation, depalmitoylation and to be sent back to the Golgi. However, unlike PDE6δ transport that shuttles a single Ras molecule, rasosomes can spread tens of Ras molecules simultaneously, enhancing the robustness of signal transduction.23 Therefore, this model suggests that rasosomes participate in spreading the signals of active Ras in the cell, providing Ras the ability to signal from various cell organelles.10
A model based on recent experiments18 and on the new data presented here is drawn in Figure 5; and describes the multiple steps involved in N-Ras or monopalmitoylated H-Ras trafficking.
Schematic model: Rasosomes bud from the Golgi to facilitate Ras trafficking and signaling. Trafficking of H-Ras and N-Ras involves palmitoylation in the Golgi, formation of Ras-loaded rasosomes that bud in a directed motion from the Golgi followed by randomized movement through the cytoplasm towards the PM. The palmitoylated and farnesylated Ras can then be exchanged and loaded on the PM, where it undergoes depalmitoylation by the palmitoyl thioesterase APT1. The depalmitoylated Ras is dislodged from the PM by PDE6δ and this complex diffuses towards the Golgi while interacting with Arl2/3-GTP that induces disintegration of the complex and loading of the depalmitoylated Ras onto the Golgi. The singly lipidated Ras is then repalmitoylated by the protein acyltransferase PAT, and can form new rasosomes
The trafficking and function of the fully processed dual palmitoylated H-Ras does not seem to fit to this scheme because neither PDE6δ18 nor rasosomes (Figure 1) are efficient interactors with H-Ras. Therefore, we suggest that the signaling of H-Ras is enhanced differently. Galectin-1 strengthens the binding of the active H-Ras-GTP form to the PM thus induces H-Ras nanoclustering and robust signaling to ERK. Therefore, the low efficiency transport of the double palmitoylated H-Ras on rasosomoes may be compensated by the trapping of active H-Ras-GTP by Gal-1. Determining the conditions at which each of the various Ras chaperones operate and how this contributes to normal Ras signaling and to its malfunctions may help in the design of novel drugs for cancer therapy.
Materials and Methods
Cell culture
COS-7 cells and MEFs were maintained in DMEM containing 10% fetal calf serum, 2 mM L-glutamine, and 100 U/ml penicillin–streptomycin (Biological Industries, Beit Haemek, Israel), in a humidified atmosphere with 5% CO2 at 37 °C. Transgenic Gal-1 knockout (Gal-1−/−), Gal-3 knockout (Gal-3−/−), and Gal-1/Gal-3 double knockout (Gal-1/3−/−, DKO) MEF cells were prepared from transgenic mice as described previously.27
Plasmids and transfection procedures
The expression constructs for GFP-H-Ras-WT, GFP-H-Ras-12V, GFP-N-Ras-WT, GFP-N-Ras-G13V, mRFP-H-Ras, and GFP-H-Ras-S17N have been described previously.19, 21, 23, 25, 32, 43 GFP-H-Ras-C181S and GFP-H-Ras-C184S were constructed using PCR site-directed mutagenesis with Phusion DNA polymerase (Finnzyme, Vantaa, Finland). For the GFP-H-Ras-C181S construct, GFP-H-Ras-WT was used as a template with the following primers: 5′-AGAGTGGCCCCGGCAGCATGAGCTGCAAG-3′ and 5′-CTTGCAGCTCATGCTGCCGGGGCCACTCT-3′.
For the GFP-H-Ras-C184S construct, the same template was used with the following primers: 5′-CGGCTGCATGAGCTGCAAGTGTGTGCTCT-3′ and 5′-AGAGCACACACTTGCAGCTCATGCAGCCG-3′.
The GFP-H-Ras-12V-C181S and GFP-H-Ras-12V-C184S mutants were generated using the same procedure but using GFP-H-Ras-12V as a template. The plasmid encoding PA GFP-H-Ras-WT, was a kind gift from Professor Mark Philips (NYU). The plasmid encoding PA-GFP-N-Ras-WT was created by inserting PCR-amplified N-Ras-WT into PA-GFP-C1 using the EcoRI and BamHI sites. The plasmid encoding PA-GFP-H-Ras-C181S was generated with the same enzymes and primers as the plasmid encoding GFP-H-Ras-C181S but the template was PA-GFP-H-Ras-WT. All mutations were verified by sequencing. Transfections were done 24 h after cell plating, using JetPEI (Plyplus-transfection, lllkirch, France) according to the manufacturer’s instructions.
Live cell imaging
Cells plated on glass cover slips were transfected 24 h after plating with plasmids encoding fluorescently tagged proteins, and imaged 21–24 h after transfection. During imaging, cells were maintained in an imaging medium containing 10 mM HEPES (pH 7.4). The imaging setup consisted of an iMIC inverted microscope with an oil-immersion Plan-Apochromatic × 100 objective NA=1.45 (Olympus, Tokyo, Japan); Polychrome V system (TILL photonics, Gräfelfing, Germany); 49 nm 100 mW DPSS laser ‘Calypso’ (Cobolt, Solna, Sweden); 561 nm 75 mW DPSS laser ‘Jive’ (Cobolt); 405 nm 60 mW diode laser iBeam (Toptica, Gräfelfing, Germany); A polytrope condenser (iMIC Beam-switch for widefield, TIRF, FRAP illumination); and an ANDOR iXon DU 888D EMCCD camera (Andor, Belfast, Northern Ireland). The microscope was surrounded by a 37 °C temperature-controlled chamber. The equipment was controlled by Live Acquisition Software (TILL photonics). Except where otherwise indicated, images were obtained every 112 ms at an exposure of 50 ms.
FRAP recordings
For photobleaching experiments, a region of interest (ROI) was marked in the cell using the Live Acquisition Software, baseline images were taken, and then GFP/RFP proteins were bleached with the 491/561 nm laser, respectively. After the bleach, the ROI was imaged as described above and its recovery was tracked. The intensity of the bleach is determined by two parameters: laser intensity and bleaching time; these parameters were always kept between 50–70% for the laser intensity, and 200–700 ms for the bleaching time.
Photoactivated GFP recordings
PA GFP is a variant of GFP that enables the user to follow a specific population of the protein of interest. PA-GFP is initially expressed in the cell in its inactive form, and can be activated by a light pulse at wavelength of 405 nm. The inactive GFP undergoes a conformational change and becomes active like standard GFP.44 In these experiments we plated the cells, transfected them with PA-GFP-tagged Ras proteins, and imaged them as described under ‘FRAP recordings’, but instead of bleaching with the 491/561 nm lasers, we used the 405 nm laser to activate the GFP. In these experiments, the amount of GFP that is activated within a given ROI is proportional to the laser intensity and the activation time; nevertheless ‘overactivation’ would cause bleaching of the GFP. Therefore, these two parameters were customized for each experiment, depending on the ROI size and the desired activation level. The values of these two parameters were kept within the limits of 30–60% for the laser intensity and 300–700 ms for the activation time.
To determine whether Ras present on the rasosomes undergoes exchange with other Ras proteins within the cell, we developed an experimental procedure using PA-GFP-Ras. Co-transfected COS-7 cells expressing PA-GFP-N-Ras-WT and mRFP-H-Ras-WT were imaged 22–26 h after transfection. The mRFP fluorescence was used to detect cells and organelles within the cell. We then performed two types of photoactivation of PA-GFP-N-Ras-WT: we photoactivated either a small part (about 20%) of the cytoplasm or the entire cell. After both activations, we recorded short time-lapse movies at t=0, t=5, t=15, and t=30 min. Using our MF algorithm,24 we calculated the number and mobility of the rasosomes and observed the decay of these factors during time.
Image processing
Tracking and finding the path of a single rasosome and measuring its motion properties in the time-lapse recordings was done using the Trackinggui algorithm as described previously.23 Calculations for rasosome quantification were done using the MF algorithm24 incorporating both the X-per index, which is a measure of the number of pixels in which a motion was observed during the movie, and the MF, which is a measure that averages the intensity of motion over all the pixels of the movie.24 Fluorescence intensities of specific cell regions were measured by a procedure in which an average intensity of all the pixels in the ROI was calculated for each frame, and then was averaged again over all of the frames in the stack, using the ROI plug-in of the WCIF ImageJ software (Rasband, W.S., ImageJ, National Institutes of Health, Bethesda, MD, USA). For dual-wavelength imaging, both wavelengths were recorded simultaneously by the Live Acquisition Software, and the two separate stacks were overlaid using MetaMorph software (Molecular Devices, Sunnyvale, CA, USA).
Western blot analysis
Western blot experiments were performed according to the standard procedures. Briefly, COS-7 cells were transfected with GFP-H-Ras-WT or with GFP-H-Ras-C181S. The cells were then lysed in 80–150 μl homogenization buffer (50 mM Tris-HCl pH 7.6, 20 mM MgCl2, 200 mM NaCl, 0.5% NP40, 1 mM DTT, and protease inhibitors). Equal amounts of proteins (20 μg per lane) were subjected to SDS-polyacrylamide (8%) gel electrophoresis. The membrane was incubated with the following antibodies: mouse anti-pan-Ras (Calbiochem, San Diego, CA, USA), rabbit anti-p-AKT (Cell Signaling, Danvers, MA, USA), mouse anti-p-ERK (Sigma, St. Louis, MO, USA), rabbit anti-AKT (Cell Signaling), rabbit anti-ERK (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and rabbit anti-β tubulin (Sigma-Aldrich). Blots were exposed to the appropriate secondary peroxidase-coupled IgG (1 : 2500) and subjected to enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Protein bands were quantified by densitometry with Image EZQuant-Gel Statistical Analysis software (EZQuant Ltd., Tel-aviv, Israel).
Abbreviations
- ATP:
-
adenosine triphosphate
- cGMP:
-
cyclic guanosine monophosphate
- DKO:
-
double knockout
- DN:
-
dominant negative
- EGF:
-
epidermal growth factor
- epi:
-
epifluorescence
- ER:
-
endoplasmic reticulum
- FRAP:
-
fluorescence recovery after photo bleaching
- Gal-1:
-
Galectin-1
- Gal-3:
-
Galectin-3
- GDI:
-
guanosine nucleotide dissociation inhibitors
- GDP:
-
guanosine diphosphate
- GFP:
-
green fluorescent protein
- GTP:
-
guanosine triphosphate
- MEF:
-
mouse embryonic fibroblast
- MF:
-
mobility factor
- mRFP:
-
monomeric red fluorescent protein
- PA:
-
photo activated
- PDEδ:
-
phosphodiesterase δ
- p-ERK:
-
phospho ERK
- PM:
-
plasma membrane
- ROI:
-
region of interest
- SE:
-
standard error
- TIRF:
-
total internal reflection fluorescence
- WT:
-
wild type
References
Malumbres M, Barbacid M . RAS oncogenes: the first 30 years. Nat Rev 2003; 3: 459–465.
Cox AD, Der CJ . The dark side of Ras: regulation of apoptosis. Oncogene 2003; 22: 8999–9006.
Downward J . Targeting RAS signalling pathways in cancer therapy. Nat Rev 2003; 3: 11–22.
Mitin N, Rossman KL, Der CJ . Signaling interplay in Ras superfamily function. Curr Biol 2005; 15: R563–R574.
Cox AD, Der CJ . Ras history: the saga continues. Small GTPases 2010; 1: 2–27.
Shields JM, Pruitt K, McFall A, Shaub A, Der CJ . Understanding Ras: '’it ain’t over ‘til it’s over’. Trends Cell Biol 2000; 10: 147–154.
Cox AD, Der CJ . Ras family signaling: therapeutic targeting. Cancer Biol Therapy 2002; 1: 599–606.
Bos JL . ras oncogenes in human cancer: a review. Cancer Res 1989; 49: 4682–4689.
Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, Cox AD, Philips MR . Ras signalling on the endoplasmic reticulum and the Golgi. Nature Cell Biol 2002; 4: 343–350.
Philips MR . Compartmentalized signalling of Ras. Biochem Soc Trans 2005; 33 (Pt 4): 657–661.
Clarke S, Vogel JP, Deschenes RJ, Stock J . Posttranslational modification of the Ha-ras oncogene protein: evidence for a third class of protein carboxyl methyltransferases. Proc Natl Acad Sci USA 1988; 85: 4643–4647.
Hancock JF, Magee AI, Childs JE, Marshall CJ . All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 1989; 57: 1167–1177.
Hancock JF, Paterson H, Marshall CJ . A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 1990; 63: 133–139.
Resh MD . Regulation of cellular signalling by fatty acid acylation and prenylation of signal transduction proteins. Cell Signal 1996; 8: 403–412.
Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK . Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol 2005; 170: 261–272.
Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI . An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science (New York, NY) 2005; 307: 1746–1752.
Zheng H, McKay J, Buss JE . H-Ras does not need COP I- or COP II-dependent vesicular transport to reach the plasma membrane. J Biol Chem 2007; 282: 25760–25768.
Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, Wittinghofer A, Bastiaens PI . The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol 2012; 14: 148–158.
Elad-Sfadia G, Haklai R, Balan E, Kloog Y . Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J Biol Chem 2004; 279: 34922–34930.
Elad-Sfadia G, Haklai R, Ballan E, Gabius HJ, Kloog Y . Galectin-1 augments Ras activation and diverts Ras signals to Raf-1 at the expense of phosphoinositide 3-kinase. J Biol Chem 2002; 277: 37169–37175.
Paz A, Haklai R, Elad-Sfadia G, Ballan E, Kloog Y . Galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage and cell transformation. Oncogene 2001; 20: 7486–7493.
Farin K, Schokoroy S, Haklai R, Cohen-Or I, Elad-Sfadia G, Reyes-Reyes ME, Bates PJ, Cox AD, Kloog Y, Pinkas-Kramarski R . Oncogenic synergism between ErbB1, nucleolin, and mutant Ras. Cancer Res 2011; 71: 2140–2151.
Rotblat B, Yizhar O, Haklai R, Ashery U, Kloog Y . Ras and its signals diffuse through the cell on randomly moving nanoparticles. Cancer Res 2006; 66: 1974–1981.
Kofer-Geles M, Gottfried I, Haklai R, Elad-Zefadia G, Kloog Y, Ashery U . Rasosomes spread Ras signals from plasma membrane 'hotspots'. Biochim Biophys Acta 2009; 1793: 1691–1702.
Belanis L, Plowman SJ, Rotblat B, Hancock JF, Kloog Y . Galectin-1 is a novel structural component and a major regulator of h-ras nanoclusters. Mol Biol Cell 2008; 19: 1404–1414.
Rotblat B, Belanis L, Liang H, Haklai R, Elad-Zefadia G, Hancock JF, Kloog Y, Plowman SJ . . H-Ras nanocluster stability regulates the magnitude of MAPK signal output. PLoS ONE 2010; 5: e11991.
Levy R, Biran A, Poirier F, Raz A, Kloog Y . Galectin-3 mediates cross-talk between K-Ras and Let-7c tumor suppressor microRNA. PloS One 2011; 6: e27490.
Shalom-Feuerstein R, Plowman SJ, Rotblat B, Ariotti N, Tian T, Hancock JF, Kloog Y . K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Res 2008; 68: 6608–6616.
Ashery U, Yizhar O, Rotblat B, Elad-Sfadia G, Barkan B, Haklai R, Kloog Y . Spatiotemporal organization of Ras signaling: rasosomes and the galectin switch. Cell Mol Neurobiol 2006; 26 (4-6): 471–495.
Ashery U, Yizhar O, Rotblat B, Kloog Y . Nonconventional trafficking of Ras associated with Ras signal organization. Traffic (Copenhagen, Denmark) 2006; 7: 119–126.
Eisenberg S, Beckett AJ, Prior IA, Dekker FJ, Hedberg C, Waldmann H, Ehrlich M, Henis YI . Raft protein clustering alters N-Ras membrane interactions and activation pattern. Mol Cell Biol 2011; 31: 3938–3952.
Niv H, Gutman O, Kloog Y, Henis YI . Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J Cell Biol 2002; 157: 865–872.
Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF . GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol 2001; 3: 368–375.
Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, Hancock JF . Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol Cell Biol 2004; 24: 6799–6810.
Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, Parton RG, Henis YI, Kloog Y, Hancock JF . Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol Cell Biol 2005; 25: 6722–6733.
Hancock JF . Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol 2003; 4: 373–384.
Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF . H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol 2000; 20: 2475–2487.
Dong X, Mitchell DA, Lobo S, Zhao L, Bartels DJ, Deschenes RJ . Palmitoylation and plasma membrane localization of Ras2p by a nonclassical trafficking pathway in Saccharomyces cerevisiae. Mol Cell Biol 2003; 23: 6574–6584.
Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR . Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 1999; 98: 69–80.
Cox AD . Protein localization: can too much lipid glue stop Ras? Nat Chem Biol 2010; 6: 483–485.
Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, Rusch M, Kramer JW, Rauh D, Coates GW, Brunsveld L, Bastiaens PI, Waldmann H . Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol 2010; 6: 449–456.
Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, Waldmann H, Bastiaens PI . The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 2010; 141: 458–471.
Goldberg L, Kloog Y. A . Ras inhibitor tilts the balance between Rac and Rho and blocks phosphatidylinositol 3-kinase-dependent glioblastoma cell migration. Cancer Res 2006; 66: 11709–11717.
Patterson GH, Lippincott-Schwartz J . A photoactivatable GFP for selective photolabeling of proteins and cells. Science (New York, NY) 2002; 297: 1873–1877.
Acknowledgements
This work was supported by Grant 2005344 from the United States-Israel Binational Science Foundation (Y. Kloog and A.D. Cox), by the Israel Science Foundation (Grant no. 912/06 to Y.K. and U.A.; Grant no. 1211/07 to U.A.), the BSF (Grant no. 2009279 to U.A.) and by the Prajs-Drimmer Institute for The Development of Anti-degenerative Drugs (Y. Kloog) and by the Tel Aviv University Center for Nanoscience and Nanotechnology (I.G.). Y. Kloog is an incumbent of The Jack H. Skirball Chair in Applied Neurobiology.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Melino
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Grunwald, A., Gottfried, I., Cox, A. et al. Rasosomes originate from the Golgi to dispense Ras signals. Cell Death Dis 4, e496 (2013). https://doi.org/10.1038/cddis.2013.16
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.16