Abstract
Autophagy is a catabolic process used to deliver cellular material to the lysosome for degradation. The core Vps34/class III phosphatidylinositol 3-kinase (PI3K) complex, consisting of Atg6, Vps15, and Vps34, is highly conserved throughout evolution, critical for recruiting autophagy-related proteins to the preautophagosomal structure and for other vesicular trafficking processes, including vacuolar protein sorting. Atg6 and Vps34 have been well characterized, but the Vps15 kinase remains poorly characterized with most studies focusing on nutrient deprivation-induced autophagy. Here, we investigate the function of Vps15 in different cellular contexts and find that it is necessary for both stress-induced and developmentally programmed autophagy in various tissues in Drosophila melanogaster. Vps15 is required for autophagy that is induced by multiple forms of stress, including nutrient deprivation, hypoxia, and oxidative stress. Furthermore, autophagy that is triggered by physiological stimuli during development in the fat body, intestine, and salivary gland also require the function of Vps15. In addition, we show that Vps15 is necessary for efficient salivary gland protein secretion. These data illustrate the broad importance of Vps15 in multiple forms of autophagy in different animal cells, and also highlight the pleiotropic function of this kinase in multiple vesicle-trafficking pathways.
Similar content being viewed by others
Main
Autophagy is an evolutionarily conserved process in which cytoplasmic proteins or organelles are packaged into lysosomes for degradation. This process can be initiated by a variety of stimuli, such as high levels of starvation or stress, to provide nutrients to the cells or to clear the cell of damaged organelles or protein aggregates.1 In some circumstances, autophagy can promote an alternative form of cell death, such as in the clearance of larval tissues in Drosophila melanogaster.2 As defects in autophagy have been implicated in several physiological and pathological conditions, such as cancer, neurodegenerative diseases, and aging,3,4 it is important to obtain a complete understanding of the molecular mechanisms controlling autophagy.
The induction of autophagy is regulated by the Atg1/Ulk1 complex, and this complex is regulated by mechanistic target of rapamycin (mTOR).5 Vesicle nucleation is controlled by the class III phosphoinositide 3-kinase (PI3K) complex that generates phosphatidylinositol 3-phosphate (PI3P).6 This conserved complex consists of vacuolar protein sorting 34 (Vps34; also known as Pik3c3), Atg6/Becn1 (also known as Vps30 in yeast), and the serine-threonine kinase Vps15/ird1 (p150 in mammals; also known as Pik3r4).7,8 Localized production of PI3P by Vps34 can act to recruit proteins containing PX or FYVE domains to membrane compartments, such as the autophagosome isolation membrane.9 Vps34 is also required more broadly for several vesicular trafficking processes such as the sorting of hydrolytic enzymes to the yeast vacuole and mammalian lysosome, and endocytic trafficking.10, 11, 12 There is mounting evidence demonstrating the pleiotropic function of the PI3K/Vps34 complex, but this has not been well studied in the context of autophagy under different physiological and cell contexts in animals.
Of the three core PI3K complex proteins, Vps15 remains an understudied kinase, and its function has not been rigorously investigated in multicellular organisms in vivo. Most of the focus on the role of this complex in autophagy regulation has been on nutrient deprivation-initiated autophagy. Indeed, previous studies determined Vps15 to be necessary for starvation-induced autophagy in the Drosophila larval fat body.13,14 However, its role in hormone-regulated autophagy, a process that occurs in the intestine,15 salivary glands,16 and fat body17 of developing Drosophila, as well as its role in other stress-induced conditions have not yet been examined. In order to address the role of Vps15 in these and other processes regulated by autophagy, we utilized Vps15 knockdown as well as a previously described null mutant14 to examine its role in a multicellular organism in vivo. We found that Vps15 is required not only for stress-induced autophagy in multiple tissues, but it is also a broad regulator of developmentally programmed autophagy in Drosophila. In addition, Vps15 is necessary for efficient protein secretion, as indicated by its role in the secretion of glue proteins from the Drosophila salivary gland. Together, these results highlight the importance of Vps15 in multiple processes in vivo.
Results
Vps15 is required for stress-induced autophagy
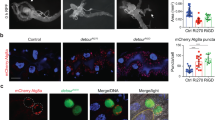
The Drosophila larval fat body undergoes a robust induction of autophagy in response to nutrient deprivation,18 and Vps15 is necessary for this autophagy.13,14 Consistent with these studies, starvation-induced formation of mCherry-Atg8a punctae, indicative of autophagosome formation, was inhibited in Vps15-null mutant cells (lacking green fluorescent protein (GFP); Figures 1a–a'') in feeding third-instar larvae starved for 4 h on 20% sucrose. We also expressed a double-stranded inverse-repeat (IR) construct that targets and knocks down expression of Vps15 (Vps15IR; Supplementary Figure S1A) in clones of cells marked with GFP. Importantly, Vps15 knockdown phenocopies the results found with the Vps15 mutant, with GFP-positive knockdown cells lacking mCherry-Atg8 punctae, whereas their wild-type neighboring cells (lacking GFP) have a robust autophagic response (Figures 1b–b''). Similar results were obtained using a different Vps15 RNAi line (Supplementary Figure S2). Consistent with these results, pharmacological inhibition of mTOR signaling by feeding Drosophila larvae rapamycin, mimicking a starvation-like signal, increases autophagosome formation in the gut, and Atg8a puncta formation is blocked by Vps15 knockdown (Figures 1c–c'').
Vps15 is required for starvation and rapamycin-induced autophagy in Drosophila. (a–a'') Fat body was dissected from feeding third-instar larvae starved for 4 h by feeding 20% sucrose. mCherry-Atg8a punctae reflect starvation-induced autophagosome formation in control (GFP positive) larval fat body cells, whereas the lack of puncta formation reflects a defect in autophagy in ΔVps15 (GFP negative) fat body cells. (b–b'') Knockdown of Vps15 phenocopies the results seen in Vps15 mutant cells as Vps15IR-expressing cells (GFP positive) lack mCherry-Atg8a punctae. (c–c'') Midguts dissected from rapamycin-fed third-instar larvae expressing Vps15IR specifically in GFP-marked clones of cells and analyzed by fluorescence and DIC microscopy. Representative images are shown
We next sought to determine whether Vps15 functions broadly in multiple forms of stress-induced autophagy. Because larvae exposed to hypoxia leave their food and cease feeding,19 it was impossible to separate the hypoxia-induced autophagic response from the starvation-induced response in vivo. Because of this, we used the macrophage-like Drosophila S2R+ cells to assess the impact of Vps15 on hypoxia-induced autophagy, as they are known to mount an autophagic response upon acute exposure to a hypoxic environment.20 In order to test the role of Vps15 in this process, S2R+ cells were transfected with the pGFP-Atg8a plasmid that expresses Atg8a fused to GFP from the endogenous Atg8a promoter.21 Exposure of these cells to a hypoxic environment (0.5% O2) leads to a large increase in Atg8a puncta formation (Figure 2a), which is also seen when the cells are soaked with control double-stranded RNA (dsRNA) targeting firefly luciferase (Figure 2a′). However, when the cells are treated with dsRNA targeting Vps15 (Supplementary Figure S1B) prior to hypoxia exposure, very few GFP-Atg8a punctae form (Figure 2a''). These results indicate that Vps15 is necessary for hypoxia-induced autophagy in Drosophila cells.
Vps15 is required for stress-induced autophagy in various tissues. (a–a'') Drosophila pGFP-Atg8a S2R+ cells were pretreated with water (a) or dsRNAs targeting firefly luciferase (luc) or Drosophila Vps15 (a' and a'') and were then placed in a hypoxic (0.5% O2) environment for 24 h prior to imaging. Knockdown of Vps15 blocked GFP-Atg8a puncta formation (a''). (b–b'') Midgut cells from third-instar larvae fed on standard food plus water (control) or 1.5% H2O2 (c–c'') for 7 h. H2O2 treatment induced the appearance of numerous mCherry-Atg8a-positive large autophagosomes (c'–c''). Vps15IR cells (GFP positive) lack mCherry-Atg8a punctae, indicating a lack of autophagy in these cells even under oxidative stress. Representative images are shown
The Drosophila larval intestinal epithelium is also sensitive to environmental perturbations, inducing autophagy in response to various stressors such as oxidative stress.22 It was previously shown that exposure of early third-instar larvae, a stage prior to the onset of developmental autophagy, to food supplemented with 1.5% H2O2 induces autophagy in intestinal cells.22 Indeed, larvae given food supplemented with only water as a control did not experience an induction of autophagy (Figures 2b–b''), whereas food supplemented with 1.5% H2O2 induced the formation of multiple autophagosomes throughout the intestine, as visualized by mCherry-Atg8a puncta formation. Importantly, autophagy induction was blocked in Vps15 knockdown cells (GFP positive; Figures 2c–c''), whereas neighboring wild-type cells (GFP negative) exhibited a robust autophagic response. Thus, it appears that Vps15 has a broad role in the regulation of stress-induced autophagy in vivo, as it is necessary for autophagy induced by starvation, hypoxia, as well as oxidative stress in multiple tissues or cell types.
Vps15 is required for developmentally programmed cell size reduction and autophagy
In addition to its important role in cell survival under conditions of stress, autophagy is required for cell death in several Drosophila tissues during development.21,23, 24, 25 The length of the Drosophila midgut decreases drastically in the dying larval intestine at the onset of puparium formation, and this structural change relies on an autophagy-dependent programmed cell size reduction.26 To determine whether Vps15 is required for programmed cell size reduction, we clonally knocked down the function of Vps15 and examined size in the midgut of animals 2 h after puparium formation (APF). Unlike their neighboring control cells that decreased in cell size in the pupal transition, Vps15IR-expressing cells remained large (Figures 3a–a'') and were found to be significantly larger compared with their neighboring control cells (Figure 3b). Consistent with these results, knockdown of Vps15 by expression of Vps15IR in all of the larval intestine cells inhibits the degradation of the midgut, as indicated by the significant inhibition of midgut area reduction (Supplementary Figure S3). These data indicate that Vps15 is required for proper developmentally programmed cell and intestine size reduction.
Vps15 is required for developmentally programmed cell size reduction and autophagy in the Drosophila midgut. (a–a'') Midguts dissected from animals expressing Vps15IR specifically in GFP-marked clones of cells 2 h APF and analyzed by fluorescence and DIC microscopy. Representative images are shown. (b) Quantification (μm2) from n=3 animal intestines with 3–7 cells measured per intestine. (c–c'') Midguts dissected from animals 0 h APF that contain a ΔVps15 mutant cell clone (lacking GFP) and analyzed by fluorescence microscopy. Wild-type (+/+) control cells possess stronger GFP and heterozygous ΔVps15/wild-type (−/+) cells have weaker GFP. Representative images are shown. (d) Quantification from n=4 animal intestines/genotype with 1–2 cells measured per intestine. (e–e'') Midguts expressing mCherry-Atg8a in all cells and expressing Vps15IR specifically in GFP-marked cell clones dissected at 2 h APF. Representative images are shown. (f) Quantification from n=3 intestines with two cells measured per intestine. Quantification is shown as mean±S.D.
The reduction in midgut cell size is accompanied by the induction of autophagy. Thus, the midgut was examined at puparium formation (0 h APF), a stage of development in which autophagy is known to occur.26 Although this developmental autophagy was marked by Atg8a puncta formation in wild-type cells (GFP positive), Atg8a localization in ΔVps15 (GFP negative; Figures 3c–c'') cells was diffuse, indicating a lack of autophagy induction. Similar results were seen with Vps15 knockdown in midguts 2 h APF where Vps15IR cells (GFP positive) lacked punctae and their neighboring wild-type cells (GFP negative) exhibited a robust induction of autophagy (Figures 3e–e'').
Steroid-activated programmed cell death of Drosophila salivary glands also requires autophagy in addition to the activation of caspases.23 The larval salivary glands of Drosophila undergo programmed cell death 14–16 h after puparium formation, resulting in no visible gland remnants by 24 h after puparium formation. However, when autophagy is blocked, salivary gland degradation is incomplete. Histological analyses of salivary glands 24 h APF, a stage at which all gland fragments should be cleared during normal development and in control animals (Figure 4a), revealed that salivary gland fragments remain in animals expressing Vps15IR using the salivary gland-specific driver, fkh-GAL4 (Figure 4a'). All pupae expressing Vps15IR in salivary glands possessed persistent salivary gland material (n=16), whereas only 8% of control animals (n=13) exhibited gland fragments (Figure 4b). Similar results were also found using a different RNAi line that expresses RNAi targeting a unique region of Vps15 (Figures 4c–c') where 67% of the pupae expressing Vps15IR in salivary glands (n=27) had persistent salivary gland cell fragments, whereas only 7% of control animals (n=15) exhibited gland cell fragments (Figure 4d).
Vps15 is required for developmentally programmed salivary gland degradation. (a and a') Control animals lacking the GAL4 driver (Vps15IR/+), n=13, and those with salivary gland-specific knockdown of Vps15 (fkh-GAL4; Vps15IR), n=16, were analyzed by histology for the presence of salivary gland fragments 24 h APF. Left: original image; right: image highlighting salivary gland fragments. (b) Quantification of data from a and a' where 8% of control animals and 100% of experimental animals possess salivary gland fragments. Statistical significance was determined using a chi-squared test. (c and c') Control animals lacking the GAL4 driver (Vps15IR/+), n=15, and those with salivary gland-specific knockdown of Vps15 (fkh-GAL4; Vps15IR #2), n=27, were analyzed by histology for the presence of salivary gland fragments 24 h APF. Left: original image; right: image highlighting salivary gland fragments. (d) Quantification of data from a and a' where 7% of control animals and 67% of experimental animals possess salivary gland fragments. Statistical significance was determined using a chi-squared test
Autophagy is robustly induced by the steroid ecdysone in the fat body of animals 0 h APF.17 Similar to what is seen in the midgut and salivary glands, expression of Vps15IR in fat body cells (GFP positive) blocks mCherry-Atg8 puncta formation, whereas neighboring wild-type cells (GFP negative) experience a robust, developmentally programmed induction of autophagy (Figure 5). Similar results occur in late-feeding third-instar larvae (Figures 5a–a''), wandering third-instar larvae (Figures 5b–b''), and white prepupae (Figures 5c–c''), indicating that Vps15 is important for autophagy at all of these developmental stages. These data suggest that, like in stress-induced autophagy, Vps15 is a broad regulator of developmentally induced autophagy.
Vps15 is required for developmentally programmed autophagy in the fat body. Fat body expressing mCherry-Atg8a in all cells and expressing Vps15IR specifically in GFP-marked cell clones dissected from (a–a'') feeding third-instar larvae, (b–b'') wandering third-instar larvae, and (c–c'') white prepupae. Representative images are shown
Vps15 is necessary for salivary gland protein secretion
Recent work has implicated autophagy genes in protein secretion,27 and we have shown that key autophagy regulatory factors, such as Atg6, are required for steroid-induced secretion of glue proteins from the salivary gland at the end of larval development.28 In order to determine whether Vps15 is involved in this process, we utilized transgenic flies expressing a fusion of the secreted glue protein, salivary gland secretion protein 3 (Sgs3), and GFP, allowing us to monitor glue secretion in vivo. By 4 h APF, most glue protein is secreted from the salivary glands.28 Homozygous ΔVps15 mutant clone cells were produced, and salivary glands were dissected and examined 4 h APF to assay for any defects in glue secretion. These Vps15 mutant cells (mCherry negative) retained SgsΔ3-GFP, whereas neighboring control cells (mCherry positive) secreted SgsΔ3-GFP and were, thus, devoid of the GFP reporter (Figures 6a–a''). Furthermore, SgsΔ3-GFP was also retained in clones of cells knocking down Vps15 (Vps15IR; dsRed positive), whereas neighboring control cells (dsRed negative) secreted SgsΔ3-GFP (Figures 6b–b''). These data indicate that Vps15 is required for salivary gland protein secretion.
Vps15 is required for efficient salivary gland protein secretion. (a–a'') Control cells (mCherry positive) are able to secrete SgsΔ3-GFP, whereas homozygous ΔVps15 mutant cells (mCherry negative) retain SgsΔ3-GFP in the cytoplasm. (b–b'') Control cells (mCherry negative) are able to secrete SgsΔ3-GFP, whereas Vps15IR-expressing cells (mCherry positive) retain SgsΔ3-GFP in the cytoplasm. Representative images are shown
Discussion
Our data indicate important roles for Vps15 in both stress-induced and developmentally programmed autophagy, as well as in salivary gland glue protein secretion. Vps15 is an essential gene, and the ΔVps15 mutant allele is homozygous lethal, with animals dying at the early third-instar larval stage.14 In addition, Vps15-deficient mouse embryos die during the implantation period before E7.5,29 highlighting the important role of this protein in the developing organism. Although there is mounting evidence that Vps15 has a broad role in the regulation of autophagy in vivo, there has been no systematic characterization of its role therein.
Studies largely based in the yeast Saccharomyces cerevisiae elucidated the genes involved in the control of autophagy, and vesicle nucleation was found to be controlled by the class III PI3K/Vps34 core complex that regulates PI3P production. In Drosophila, this complex consists of Vps34, Atg6, and Vps15/ird1. Vps15 was first identified in a screen seeking to identify mutations that lead to defects in the localization of vacuolar hydrolases in S. cerevisiae,30 and the majority of the published work since has focused on its role in nutrient deprivation-induced autophagy. Vps15 has also been shown to be necessary for the robust starvation-induced autophagy response in mammalian cells.29 However, autophagosomes form without issue in the skeletal muscle of Vps15 muscle-specific knockout mice upon starvation. This contrasts with the reported defects of autophagy initiation in Vps34-deficient liver and heart31 but agrees with work showing that Vps34 deletion in sensory neurons leaves the autophagy pathway intact.32 Indeed, our work and others have shown that regulators of autophagy, such as Atg7, may have different regulatory mechanisms in distinct cell types within an animal and that different forms of autophagy could involve unique regulatory pathways.26,33,34 Thus, it is important to characterize the role of autophagy-related molecules according to the type of autophagy in which they are involved and in a tissue-specific manner.
In addition to its previously described role in starvation-induced autophagy in the Drosophila larval fat body, we have shown Vps15 to be necessary for other types of stress-induced autophagy in other tissues. In the S2R+ cell line derived from Drosophila blood cells, Vps15 is necessary for the cell’s ability to mount an autophagic response in conditions of acute hypoxia. This agrees with previous results showing that knockdown of Atg5, an E3 ubiqutin ligase involved in autophagosome elongation, blocks hypoxia-induced autophagy in tumor cells.20 In addition, Vps15 was shown to be necessary for autophagy induction in response to oxidative stress in the Drosophila midgut of the intestine. This agrees with previous data showing that suppression of other core autophagy genes such as Atg126 blocks H2O2-induced autophagy in the intestine. Importantly, like other genes involved in the autophagic pathway,17,23,26,35 we were also able to demonstrate the significance of Vps15 in developmentally programmed autophagy. In the Drosophila intestine, Vps15 was shown to be necessary for programmed cell size reduction and induction of autophagy. Salivary gland clearance and programmed autophagy in the fat body were also shown to be reliant on Vps15, clearly indicating a broad role for Vps15 in the regulation of autophagy in multiple cell types and contexts within an animal.
Recent studies have indicated that autophagic factors regulate protein secretion,27 and we have shown that both Atg6 and Vps34 as well as Atg1 are required for protein secretion in salivary gland cells.28 Here, we show that loss of Vps15 in salivary gland cells leads to a disruption in protein secretion, supporting previous evidence that protein secretion might be an autophagy-dependent process.28 However, as Beclin 1 and PI3P also localize to the trans-Golgi network,36,37 it is also possible that Vps15 may be acting as a part of an autophagy-independent process regulating protein secretion. Future studies should elucidate the role of autophagy factors and possibly PI3P in this process.
Materials and Methods
Fly stocks and culture
Flies were reared at 25°C on standard cornmeal/molasses/agar media. The following Drosophila stocks were used: ΔVps15 (from H Stenmark, University of Oslo and The Norwegian Radium Hospital, Oslo, Norway), P{UAS-ird1-RNAi} NIG 9746R-2 (from the National Institute of Genetics (NIG), Mishima, Shizuoka, Japan), UAS-ird1 RNAi (#34092; described as Vps15IR #2 in the text) and w; SgsΔ3-GFP (Bloomington Drosophila Stock Center, Bloomington, Indiana, USA), yw,hs-Flp; Cg-GAL4, UAS-mCherry-Atg8a; FRT82B, UAS-GFPnls (from T Neufeld, University of Minnesota, MN, USA), yw,hs-Flp; pmCherry–Atg8a; Act>CD2>GAL4, UAS–nlsGFP/TM6B, yw,hs-Flp; +; hs-GFP-Atg8a, Act>CD2>GAL4, UAS-dsRed, NP1-GAL4, and fkh-GAL4. ΔVps15 flies were recombined onto an FRT82B chromosome (Bloomington Drosophila Stock Center).
Cell culture and hypoxia treatment
Drosophila S2R+ cells were obtained from Drosophila Genomics Resource Center (DGRC, Indiana University, Bloomington, IN, USA) and were grown at 25°C in Schneider’s Drosophila medium supplemented with 10% fetal bovine serum (FBS), 1 × GlutaMAX, and penicillin-streptomycin (Gibco, Life Technologies, Carlsbad, CA, USA).
dsRNA for Vps15 RNAi treatment was produced by in vitro transcription of a polymerase chain reaction (PCR)-generated DNA template from Drosophila genomic DNA containing the T7 promoter sequence on both ends. Genomic DNA was harvested from Drosophila S2 cells using the Wizard Genomic DNA Purification Kit from Promega (Madison, WI, USA). The Photinus pyralis luciferase DNA template was amplified from the pGL3 control vector (Promega). Target sequences were scanned to exclude any complete 19-mer homology to other genes. dsRNAs were generated using the MEGAscript T7 kit (Ambion, Austin, TX, USA) and purified using the Qiagen RNeasy kit (Valencia, CA, USA). The primer sequences used for the generation of dsRNAs were as follows: luciferase: forward, 5′-taatacgactcactatagggGCTGGGCGTTAATCAGAGAG-3′, reverse, 5′-taatacgactcactatagggTTTTCCGTCATCGTCTTTCC-3′; Vps15: forward, 5′-taatacgactcactatagggAAGACGGTCCTGTTGTGGAC-3′, and reverse, 5′-taatacgactcactatagggCCGGTGAGAATAAAGGGTGA-3′.
A solution containing 20 μg dsRNA in water or an equivalent amount of water as a control was added to triplicate wells in 6 well plates. pGFP-Atg8a S2R+ cells were resuspended in serum-free media at 0.5 × 106 cells/ml and 1 ml of this suspension as a control was added to the wells containing dsRNAs. Cells were incubated at room temperature for 30 min after which 3 ml of complete medium was added. The cells were incubated for 3 additional days at 25°C and were imaged using an Olympus IX71 inverted microscope (Olympus, Shinjuku, Tokyo, Japan). Images were acquired with a QImaging Retiga 1300 camera and processed using QCapture 2.99.5 (QImaging, Burnaby, BC, Canada).
Induction of cell clones
To induce knockdown in clones of cells, virgin females of yw,hs-Flp; pmCherry–Atg8a; Act>CD2>GAL4, UAS–nlsGFP/TM6B or yw,hs-Flp; +; hs-GFP-Atg8a, Act>CD2>GAL4, UAS-dsRed were crossed to indicated RNAi or transgenic lines. One-day egg lays were heat shocked at 37°C for 15 min. To induce loss-of-function mutant cell clones, yw,hs-Flp; Cg-GAL4, UAS-mCherry-Atg8a; FRT82B, UAS-GFPnls or yw, hs-Flp; pmCherry–Atg8a; FRT82B, Ubi-nlsGFP virgins were crossed to FRT82B, ΔVps15 flies. One-day egg lays were heat shocked for 1 h at 37°C.
Analysis of knockdown efficiency
As commercial antibodies against Drosophila Vps15 were not available, knockdown efficiency was analyzed using real time PCR (RT-PCR). For in vivo experiments, 20 intestines from either male (control) or female (knockdown) wandering third-instar larvae from NP1-GAL4 males crossed to Vps15 IR (x) females or 20 intestines of female offspring from a w1118 × Vps15 IR (x) cross were dissected and placed in Schneider's Drosophila medium supplemented with 10% FBS, 1x GlutaMAX, and penicillin-streptomycin (Gibco, Life Technologies). For in cellulo experiments, cells were harvested from triplicate wells of six-well plates of pGFP-Atg8a S2R+ cells soaked with luciferase or Vps15 dsRNA as previously described and were washed with 1x phosphate-buffered saline solution (1x PBS). RNA was isolated from both the cells and intestines using the Qiagen RNeasy kit (Valencia, CA, USA) according to the manufacturer’s instructions and cDNA was produced using the SuperScript III First-Strand Synthesis System (Invitrogen, Life Technologies, Carlsbad, CA, USA).
RT-PCR was performed using the default PCR cycle on a sequence detection system (ABI Prism 7900 HT, Applied Biosystems, Life Technologies, Carlsbad, CA), and amplified cDNA was detected using SYBR green dye (Power SYBR Green PCR Master Mix, Applied Biosystems). Thermocycling conditions used for quantitative PCR were 1 cycle at 95°C for 10 min and a total of 40 cycles at 95°C for 15 s, 56°C for 30 s, and 72°C for 30 s. Quantification of relative amounts of Vps15 was performed using the Sequence Detection Systems version 2.3 software (Applied Biosystems). RT-PCR was conducted in triplicate for each sample. Two primer pairs for Vps15 (forward, 5′-AGCACTGGAGGCACGATCAC-3′, reverse, 5′-GTCCCATCTCCTCGTACTG-3′; forward, 5′-GAGATGGGACAGACCTTG-3′, reverse, 5′-GAGATAAGGAACGGGTTCATGG-3′) along with primers for the reference genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH; forward, 5′-CATTGTGGGCTCCGGCAA-3′, reverse, 5′-CGCCCACGATTTTCGCTATG-3′) and ribosomal protein 32 (RpL32; forward, 5′-AGCATACAGGCCCAAGATCG-3′, reverse, 5′-TGTTGTCGATACCCTTGGGC-3′) were used for RT-PCR and each Vps15 primer pair was separately normalized to each reference gene.
Starvation
To initiate starvation-induced autophagy, third-instar larvae were transferred from standard food to 20% sucrose in water for 4 h prior to dissection. Control larvae were transferred to standard food until dissection. The fat body was dissected in 1x PBS and was fixed briefly for about 3 min in 4% paraformaldehyde (PFA). It was then stained with Hoechst 33342 trihydrochloride, trihydrate (Invitrogen, Life Technologies, Carlsbad, CA, USA) and washed in 1x PBS before mounting in Vectashield (Vector Laboratories, Burlingame, CA, USA). Images were collected on a Zeiss AxioImager Z1 microscope equipped with an Apotome (Carl Zeiss Microscopy, Jena, Germany). Images were acquired with Axiocam and minimally processed using Zeiss Axiovision 4.9.1 (Carl Zeiss Microscopy) and Adobe Photoshop CS6 16.0.3 (Adobe, San Jose, CA, USA).
Rapamycin treatment
Early third-instar larvae were washed in PBS and starved for 45 min in 20% sucrose in PBS prior to placing on food containing either 5 μM rapamycin (Sigma-Aldrich, St. Louis, MO, USA; from 1 mM in ethanol) or an equivalent amount of ethanol only (control) for 4 h. The intestines were dissected, fixed for a minimum of 30 min in 4% PFA, washed once with 1x PBS, and once with 0.1% Triton X-100 (Sigma-Aldrich) in PBS, and were allowed to incubate overnight in Vectashield supplemented with DAPI (Vector Laboratories). The next day, the intestines were imaged using a Zeiss AxioImager Z1 microscope (Carl Zeiss Microscopy) equipped with an Apotome, as previously described.
Oxidative stress treatments
Early third-instar larvae were collected and starved in the presence of water-soaked filter paper for 45 min at 25°C prior to stress exposure. Subsequently, larvae were placed on food containing 1.5% H2O2 (Fisher Scientific, Fair Lawn, NJ, USA) or an equivalent amount of water as a control for 7 h. The intestines were dissected, fixed for a minimum of 30 min in 4% PFA, washed once with 1x PBS, and once with 0.1% Triton X-100 (Sigma-Aldrich) in PBS, and were allowed to incubate overnight in Vectashield supplemented with DAPI (Vector Laboratories). The next day, the intestines were imaged using a Zeiss AxioImager Z1 microscope (Carl Zeiss Microscopy) equipped with an Apotome, as previously described.
Quantification of cell size
Cell size was quantified using the measure outline function of Zeiss Axiovision 4.9.1 software (Carl Zeiss Microscopy).
Histology
Flies were maintained at 25°C and aged to 24 h APF. For histology, whole pupae were fixed and processed as described previously38 for paraffin sectioning and light microscopy.
Protein secretion assay
Salivary glands were dissected from control and mutant animals 4 h APF, fixed for 30 min in 4% PFA, washed briefly two times in PBS, and mounted in Vectashield with DAPI.
Quantification and statistical analyses
ImageJ (NIH, Bethesda, MD, USA) was used for the quantification of Atg8a puncta. For these experiments, Student’s t-test for two samples assuming unequal variances was used to determine the statistical significance of the data. For salivary gland fragment analysis, significance was determined using the chi-square test. Statistical analyses were performed using GraphPad Prism software (GraphPad Software, San Diego, CA, USA).
Abbreviations
- APF:
-
after puparium formation
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- GFP:
-
green fluorescent protein
- FYVE:
-
Fab1, YOTB, Vac1 and EEA1
- IR:
-
inverse repeat
- ird1:
-
immune response deficient 1
- PCR:
-
polymerase chain reaction
- PI3K:
-
phosphoinositide 3-kinase
- PI3P:
-
phosphatidylinositide 3-phosphate
- PX:
-
phox homology
- RFP:
-
red fluorescent protein
- RpL32:
-
ribosomal protein L32
- S2R+:
-
Schneider's line 2, receptor positive
- Sgs3:
-
salivary gland secretion protein 3
- TOR:
-
target of rapamycin
- Vps15:
-
vacuolar protein sorting 15
References
Mizushima N, Komatsu M . Autophagy: renovation of cells and tissues. Cell 2011; 147: 728–741.
Das G, Shravage BV, Baehrecke EH . Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb Perspect Biol 2012; 4: 321–334.
Mizushima N, Levine B, Cuervo AM, Klionsky DJ . Autophagy fights disease through cellular self-digestion. Nature 2008; 451: 1069–1075.
Levine B, Kroemer G . Autophagy in the pathogenesis of Disease. Cell 2008; 132: 27–42.
Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y . Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 2000; 150: 1507–1513.
Ohsumi Y . Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001; 2: 211–216.
Funderburk SF, Wang QJ, Yue Z . The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20: 355–362.
Kihara A, Noda T, Ishihara N, Ohsumi Y . Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 2001; 152: 519–530.
Lindmo K, Stenmark H . Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci 2006; 119: 605–614.
Brown WJ, DeWald DB, Emr SD, Plutner H, Balch WE . Role for phosphatidylinositol 3-kinase in the sorting and transport of newly synthesized lysosomal enzymes in mammalian cells. J Cell Biol 1995; 130: 781–796.
Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD . Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 1993; 260: 88–91.
Stack JH, Herman PK, Schu PV, Emr SD . A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J 1993; 12: 2195–2204.
Juhász G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM et al. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol 2008; 181: 2347–2360.
Lindmo K, Brech A, Finley KD, Gaumer S, Contamine D, Rusten TE et al. The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy 2008; 4: 500–506.
Lee C-Y, Cooksey BAK, Baehrecke EH . Steroid regulation of midgut cell death during Drosophila development. Dev Biol 2002; 250: 101–111.
Lee C-Y, Baehrecke EH . Steroid regulation of autophagic programmed cell death during development. Development 2001; 128: 1443–1455.
Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell 2004; 7: 179–192.
Scott RC, Schuldiner O, Neufeld TP . Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7: 167–178.
Wingrove JA, O'Farrell PH . Nitric oxide contributes to behavioral, cellular, and developmental responses to low oxygen in Drosophila. Cell 1999; 98: 105–114.
Wilkinson S, O'Prey J, Fricker M, Ryan KM . Hypoxia-selective macroautophagy and cell survival signaled by autocrine PDGFR activity. Genes Dev 2009; 23: 1283–1288.
Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol 2009; 19: 1741–1746.
Wu H, Wang MC, Bohmann D . JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech Dev 2009; 126: 624–637.
Berry DL, Baehrecke EH . Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 2007; 131: 1137–1148.
Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjørkøy G, Johansen T et al. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol 2010; 190: 523–531.
Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM . Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol 2008; 182: 1127–1139.
Chang T-K, Shravage BV, Hayes SD, Powers CM, Simin RT, Harper JW et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol 2013; 15: 1067–1078.
Deretic V, Jiang S, Dupont N . Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol 2012; 22: 397–406.
Shravage BV, Hill JH, Powers CM, Wu L, Baehrecke EH . Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development 2013; 140: 1321–1329.
Nemazanyy I, Blaauw B, Paolini C, Caillaud C, Protasi F, Mueller A et al. Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol Med 2013; 5: 870–890.
Robinson JS, Klionsky DJ, Banta LM, Emr SD . Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol 1988; 8: 4936–4948.
Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci USA 2012; 109: 2003–2008.
Zhou X, Wang L, Hasegawa H, Amin P, Han BX, Kaneko S et al. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci USA 2010; 107: 9424–9429.
McPhee CK, Logan MA, Freeman MR, Baehrecke EH . Activation of autophagy during cell death requires the engulfment receptor Draper. Nature 2010; 465: 1093–1096.
Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009; 461: 654–658.
Denton D, Chang TK, Nicolson S, Shravage B, Simin R, Baehrecke EH et al. Relationship between growth arrest and autophagy in midgut programmed cell death in Drosophila. Cell Death Differ 2012; 19: 1299–1307.
Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 2000; 19: 4577–4588.
Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T . Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep 2001; 2: 330–335.
Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ et al. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 2006; 133: 3305–3315.
Acknowledgements
We thank L Wu, H Stenmark, and the Bloomington Stock Center for fly strains; N Silverman and G Das for S2 cells; Kirsten Tracy for the ImageJ puncta analysis macro; Tina Fortier for technical support; and members of the Baehrecke laboratory for constructive feedback. This work was supported by NIH grants GM079431 and CA159314 to EHB. EHB is an Ellison Medical Foundation Scholar.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by G Kroemer
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Anding, A., Baehrecke, E. Vps15 is required for stress induced and developmentally triggered autophagy and salivary gland protein secretion in Drosophila. Cell Death Differ 22, 457–464 (2015). https://doi.org/10.1038/cdd.2014.174
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2014.174
This article is cited by
-
RNAi screening identifies Trypanosoma brucei stress response protein kinases required for survival in the mouse
Scientific Reports (2017)
-
Autophagy in stress and disease
Cell Death & Differentiation (2015)
-
Distinct requirements of Autophagy-related genes in programmed cell death
Cell Death & Differentiation (2015)
-
Current questions and possible controversies in autophagy
Cell Death Discovery (2015)