Abstract
Intrinsic and acquired chemoresistance are frequent causes of cancer eradication failure. Thus, long-term cis-diaminedichloroplatine(II) (CDDP) or cisplatin treatment is known to promote tumor cell resistance to apoptosis induction via multiple mechanisms involving gene expression modulation of oncogenes, tumor suppressors and blockade of pro-apoptotic mitochondrial membrane permeabilization. Here, we demonstrate that CDDP-resistant non-small lung cancer cells undergo profound remodeling of their endoplasmic reticulum (ER) proteome (>80 proteins identified by proteomics) and exhibit a dramatic overexpression of two protein disulfide isomerases, PDIA4 and PDIA6, without any alteration in ER-cytosol Ca2+ fluxes. Using pharmacological and genetic inhibition, we show that inactivation of both proteins directly stimulates CDDP-induced cell death by different cellular signaling pathways. PDIA4 inactivation restores a classical mitochondrial apoptosis pathway, while knockdown of PDIA6 favors a non-canonical cell death pathway sharing some necroptosis features. Overexpression of both proteins has also been found in lung adenocarcinoma patients, suggesting a clinical importance of these proteins in chemoresistance.
Similar content being viewed by others
Main
One of the major aims of chemotherapy is to induce apoptosis to eradicate cancer cells, but cancer cells are intrinsically resistant or acquire chemoresistance to apoptosis that may lead to treatment failure.1, 2 Thus, many chemotherapeutic agents such as the DNA-damaging agent cisplatin (cis-diaminedichloroplatine(II) (CDDP)) initiate a mitochondrial cell death pathway involving Bcl-2 family members, p53, mitochondrial membrane permeabilization (MMP), cytochrome c release and caspase activation in tumor cells.3, 4, 5 Following several years of treatment, CDDP-treated tumors, such as lung, ovarian, testicular and head and neck carcinomas, develop resistance to CDDP-induced apoptosis. Although causes of chemoresistance can be multiple, adaptation to endoplasmic reticulum (ER) stress, as a result of chronic and mild unfolded protein response (UPR), might be a key driver of malignancy and resistance to therapy.6, 7, 8, 9
The UPR is activated when misfolded proteins accumulate in the ER as a result of exogenous and/or endogenous stress signals.8 Although ER stress responses represent homeostatic mechanisms allowing cells to survive, prolonged or excessive activation of the UPR can result in cell death by inducing primarily mitochondrial apoptosis.10, 11 UPR is regulated by the balance between expression levels and post-translational modification status of ER sensor proteins, including ER to nucleus signaling 1 (IRE1), protein kinase RNA-like endoplasmic reticulum kinase (PERK) and activating transcription factor 6 (ATF6). It is frequently accompanied by an altered calcium homeostasis and autophagy.8 Moreover, 78 kDa glucose-regulated protein (GRP78) overexpression has been associated with enhanced tumor growth and resistance to chemotherapy.12, 13 However, how the UPR switches between the pro-survival and pro-apoptotic signaling pathways14, 15 and therefore how it might contribute to cancer cell resistance is still unknown.
Here we addressed the hypothesis that CDDP resistance of non-small lung cancer (NSLC) relies on specific adaptation mechanisms involving ER resident proteins such as protein disulfide isomerase (PDI) without any alteration of Ca2+ fluxes between ER and mitochondria. A set of CDDP-resistant NSLC A549 cell lines16 and lung cancer patients biopsies were investigated to identify novel anti-apoptotic proteins responsible for CDDP resistance. Accordingly, pharmacological inhibition and genetic manipulation of PDIA4 and PDIA6 restored cell death induction in CDDP-resistant clones, revealing for the first time their role in cancer cell adaptation and chemoresistance.
Results
Chronic adaptation of lung carcinoma cells to CDDP involves the alteration of the UPR pathway in the ER
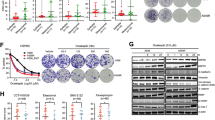
A549 lung adenocarcinoma cells (wild type, WT) were cultured in the presence of low doses of CDDP (5 μM), until the appearance of resistant cells able to survive and maintain their resistance phenotype in the absence of the selection pressure and three clones, named A549 R1, R2 and R3, were isolated.16 Using a clonogenic assay, the proliferation capacity of these clones was evaluated following culture of a similar number of cells of the resistant clones for 48 h before plating. Resistant cells appeared to grow faster than WT cells with a significant difference between each clone and WT (P<0.01; Figures 1a and b).
CDDP-resistance lung cell model. (a) A549 wild-type cells (WT) and resistant R1 to R3 clones from 4-day precultures were plated and incubated for 14 days. (b) Colony count shows that R1 grows faster than R2, R3 and WT cells. Data points are mean±S.E.M. from three experiments performed in triplicate (n=3, mean±S.E.M.). (c and d) WT and resistant clones were incubated with 0 (NT), 25 and 50 μM CDDP. (c) The level of the ΔΨm was measured with the TMRM fluorescent probe by flow cytometry at 24 h (n=6, mean±S.D.). (d) DNA content was measured with propidium iodide at 48 h (n=6, mean±S.D.). (e) Western blotting analysis of apoptotic proteins, chaperones and ER stress markers in A549 WT and R1 to R3 clones. Thirty micrograms of cellular proteins have been immunoblotted by antibodies against BCL-2, BAX, PARP1, PERK, IRE1, ATF6 and GRP78. Western blots are representative of three independent experiments
Two essential parameters of apoptosis were measured according to international criteria,17 collapse of mitochondrial transmembrane potential (ΔΨm) and hypoploidy (SubG1 peak) by flow cytometry. As expected,16, 18 WT cells showed dose-dependent loss of ΔΨm with 25 and 50 μM CDDP at 24 h, indicating that CDDP treatment induces the mitochondrial pathway of apoptosis (Figure 1c). CDDP-induced loss of ΔΨm was significantly prevented in all resistant clones, although to various extent. In addition, a large fraction of WT cells appeared as a SubG1 population at 48 h following CDDP treatment, confirming the apoptosis induction by CDDP, but no significant enhancement of this event was found in resistant clones (Figure 1d). As CDDP has no direct effect on highly purified isolated mitochondria (not shown), data from the first set of experiments indicated that resistance to CDDP-induced apoptosis of R1, R2 and R3 clones involves a mechanism upstream of MMP.
Next, we characterized expression levels of several apoptosis-related proteins by immunoblotting (Figure 1e). Indeed, we observed increased Bcl-2, Hsp70 and PARP1 expression in parallel with a decreased Bax expression in the CDDP-resistant clones. This result is in line with a previously described balance between anti- and pro-apoptotic proteins directly regulating MMP and chemoresistance.15, 19, 20, 21, 22
However, these observations did not unravel the pathways upstream of MMP, which can be involved in the development of CDDP resistance. It has been previously established that ER homeostasis can be a common extra-nuclear target of CDDP.23 Moreover, our previous work established a role of the UPR and consequent ER stress as one of the most potent endogenous inducers of MMP and cell death.11 This led us to hypothesize that alteration of the UPR might be involved in the resistance to CDDP-induced cell death. Thus, we evaluated the expression of three UPR sensors, ATF6, IRE1 and PERK, and their major partner, GPR78.8 Confirming our hypothesis, all these proximal UPR factors were differentially regulated in the CDDP-resistant clones as compared with WT. ATF6 and PERK expression was downregulated, whereas the IRE1 expression increased (Figure 1e). Surprisingly, we also observed a dramatic reduction in the expression of GRP78 in all the resistant clones (Figure 1e). These results suggested that adaptation to ER stress, as shown by the differential modulation of ER proteins levels, might be a central process to the chronically acquired chemoresistance but that there is no canonical ER stress per se.
Altered ER Ca2+ homeostasis is not required for resistance to CDDP
Ca2+ release from the ER and subsequent mitochondrial Ca2+ overload and MMP have been shown to have a crucial role in ER stress-induced cancer cell death.11, 24 Thus we investigated whether development of CDDP resistance was associated with alterations of ER Ca2+ homeostasis and Ca2+-mediated MMP. First, we expressed ERD1, a FRET-based ER-targeted recombinant Ca2+ probe25 in WT and R1 cells (i.e., the clone showing the highest level of resistance) and measured steady-state Ca2+ levels under control conditions and following 24 h treatment with CDDP (75 μM), using an acceptor bleaching method previously established in our laboratory.26 We did not find any significant differences between the steady ER Ca2+ concentrations ([Ca2+]ER) in the WT and R1 cells (Figure 2a). Moreover, we observed no ER Ca2+ depletion following CDDP treatment. Thapsigargin, an inhibitor of ER Ca2+ accumulation,27 induced a large drop in the FRET signal, validating the experimental method (Figure 2a). In line with these results, no increase in cytosolic [Ca2+] ([Ca2+]c) was found following CDDP treatment, as measured in WT cells loaded with the fluorescence Ca2+-sensitive dye Fura-2 (Figure 2b). Finally, in order to assess the contribution of cellular Ca2+ signaling to CDDP-induced cell death, we applied extracellular (EGTA, Figure 2c) and intracellular (BAPTA-AM, Figure 2d) Ca2+ chelators, and an inhibitor of Ca2+ release from the ER (2-APB, Figure 2e) and quantified cell death in WT cells following 24 h treatment with CDDP. Neither EGTA nor 2-APB had any effect on the efficiency of CDDP to induce cell death, while BAPTA-AM pretreatment led to a significant but small protection (52.3±1.57% cell death in control WT cells versus 41.4±2.62% in the presence of BAPTA-AM, 10 μM, Figure 2d). In conclusion, neither Ca2+ influx nor Ca2+ release from the ER contributed significantly to CDDP-mediated cell death, implying that alterations in ER Ca2+ homeostasis cannot explain the CDDP resistance of resistant cells. The slight inhibition of CDDP-induced cell death in BAPTA-AM-treated cells might reflect inhibition of calpains, which have been previously shown to marginally contribute to caspase activation following CDDP treatment of A549 cells.16
Ca2+ signaling is not implied in CDDP-induced cell death. (a) Steady state [Ca2+]ER was measured by ERD1 probe FRET efficiency in WT and R1 A549 cells left either untreated or after 24 h treatment with CDDP (75 μM), as described in the Materials and Methods section (see also Supplementary Figure S1). As positive control, WT cells were treated with thapsigargin in the absence of extracellular Ca2+ and the presence of 100 μM EGTA. Results shown are the mean±S.E.M. of at least three different cell preparations (*P<0.05). (b) Timelapse imaging of [Ca2+]c for 180 min following CDDP treatment. Representative traces are shown on the left panel. CDDP was applied as indicated, and fura-2 loading was tested by the addiditon of 1 μM ionomycin. On the right panel, mean±S.E.M. of background-corrected 340 nm/380 nm Fura-2 ratios are shown from at least three different cell populations following 60 min of CDDP treatment. (c–e). The effect of extracellular (EGTA, c), intracellular (BAPTA-AM, d) Ca2+ chelation and inhibition of Ca2+ release from the ER (2-APB, e) on CDDP-induced cell death. The chelators and inhibitor were applied in the culture medium at the indicated concentrations either to control cells or to cells treated with CDDP (75 μM, 24 h), and cell death was measured by determination of the ratio of PI-positive cells, as described in the Materials and Methods section. Results shown are the mean±S.E.M. of at least three different cell preparations (*P<0.05)
Proteomics identifies ER adaptations mediating CDDP resistance
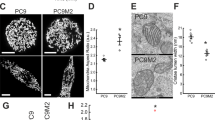
In order to identify novel ER-resident pathways contributing to CDDP resistance, we used an unbiased approach consisting of the comparison of the ER proteomes from WT and R1 cells. Proteins of an ER-enriched fraction, obtained by differential centrifugation, were separated by 2D denaturing electrophoresis. At least three replicate gels per cell type were silver-stained for quantitative analysis of the ER proteome. Among 492 ER proteins found in R1 and WT (not shown), 80 were overexpressed 2–10-fold in R1 compared with WT (Supplementary Figure S2). Forty spots were further analyzed by nanoLC/MS/MS, and 23 were identified by their MASCOT score and SwissProt accession number (Supplementary Table S1). Among this set, we identified a group of genuine ER proteins related to protein-folding functions, which belong to the PDI family. PDIA4 levels were increased 11.2-fold while PDIA6 was upregulated 7.75-fold in ER as measured by densitometry of 2D electrophoresis gels (Figure 3a). Moreover, these proteins were found to be overexpressed in total cell lysates of all CDDP-resistant clones by immunoblotting of 1D gels, except for PDIA4 whose expression remained unchanged in R3 cells (Figure 3b). These results suggested an importance of these proteins in the development of ER-mediated adaptation to CDDP-induced cellular stress. Therefore, we further explored their regulation and role.
PDIA4 and PDIA6 identification in resistant cells. (a) Semi-quantitative analysis of PDIA4 and PDIA6 amounts in WT and R1 cells. Proteins have been separated by 2D gel electrophoresis, gel has been stained by Blue Coomassie and the intensity of each spot has been determined by densitometry. RD, relative density. (b) Comparison of PDI4 and PDIA6 expression in WT and R1, R2 and R3 by immunoblotting. β-actin is used as a loading control to quantify proteins. (c) PDI activity was quantified by measuring the PDI-catalyzed reduction of insulin in the presence of DTT in 60 μg of whole extract from A549WT and A549R1 to R3. Co., buffer without PDI and hPDIr 16 μg of purified human recombinant PDI (n=3, means±S.E.M.). (d) Similarly, PDI activity was evaluated in ER fraction of WT and R1 cells and compared to controls (n=3, means±S.E.M.). (e) Stimulation of CDDP-induced ΔΨm loss by 0.5 or 1 mM Bacitracin in 48 h of treatments (n=6, means±S.E.M.)
PDI proteins are soluble Ca2+-binding chaperones containing thioredoxin-like domains,28 but little information is available on physiological PDIA4 and PDIA6 activity. Therefore, we determined whether changes in PDIA4 and PDIA6 protein levels lead to a change in cellular PDI enzymatic activity. Using an in vitro assay of insulin disulfide bonds reduction, we identified an enhancement of PDI activity in all resistant clones compared with WT cells (Figure 3c). Total PDI activity was increased in R1, R2 and R3 cells by 39, 37 and 42% of the WT, respectively. Next, we treated the resistant cells with bacitracin, a pan inhibitor of PDIs, and observed an increase in the CDDP-induced loss of ΔΨm, restoring almost completely CDDP sensitivity for the highest dose of PDI inhibitor (Figure 3d).29 These results indicate that pharmacological PDI inhibition rescues MMP induction by CDDP and suggests that PDIs might have a direct role in CDDP resistance.
Genetic downregulation of some PDI isoforms reverses CDDP resistance
Twenty-one genes are known to encode PDI family members. With the aim of identifying PDI isoform(s) responsible for CDDP resistance, we transfected pools of siRNAs to selectively and individually inhibit the expression of various PDI isoforms. Four isoforms have been selected because of their previously characterized role in survival to ER stress (PDIA1),30 in Ca2+ exchange (PDIA3)31 and their overexpression in the present study (PDIA4 and PDIA6). Thus, siRNAs against these four isoforms allowed efficient knockdown in A549-resistant clones with limited off-target effects in 48 h (Supplementary Figure S3). We compared the impact of the depletion of PDIA1, PDIA3, PDIA4 or PDIA6 on cell viability (Figure 4a). Knockdown of PDIA4 and PDIA6 increased the CDDP-induced cell death. In contrast, depletion of PDIA1 and PDIA3 had no effect on cell viability (Figure 4a). These results point to a potential direct role of PDIA4 and PDIA6 isoforms in CDDP resistance but exclude a role for PDIA1 and PDIA3. Co-targeting of both proteins was lethal without CDDP treatment, indicating that the presence of at least one of the isoforms is essential for normal cell physiology and that they might have overlapping functions (Figure 4b).
Short-term and long-term effects of PDI knockdown on CDDP treatments. (a) siRNA silencing of PDIs in R1 to R3 cells. Resistant cells were transfected for 48 h with siRNA Crtl, PDIA1, PDIA3, PDIA4 and PDIA6, and cell viability was evaluated after CDDP treatment for 48 h by WST-1 assay (n=3, mean±S.E.M.). Non-targeting siRNA, Crtl. (b) Co-silencing of PDIA4 and PDIA6 in R1 to R3 cells (n=3, mean±S.E.M.). (c) Clonogenic assay of WT and R1 from 4-day precultures in the presence of siRNA Crtl, PDIA4 and PDIA6 for 48 h, and 25 μM CDDP were plated in drug-free medium in six-well plates, incubated for 14 days and colony count was determined. (d) Flow cytometry analysis of R1 cells treated by siRNAs and CDDP, as above, labeled with 7-AAD and Annexin V. Percentage of cells are indicated in parts of the panels (n=6)
Next, we performed a clonogenic assay to examine the impact of PDI levels on the sensitivity of the clones to CDDP (Figure 4c). Addition of 25 μM CDDP abolished the clonogenic potential of WT cells but only reduced the number of R1 colonies by >2-fold, in line with previous results. After knockdown of PDIA4 or PDIA6 in R1 cells, CDDP became more efficient and a rather limited number of colonies (≤8±1) was observed, indicating restoration of CDDP-induced cell death. Similar results were obtained for R2 and R3 (not shown).
Using AnnexinV–PE/7AAD labeling procedure, we showed an increase in early and late apoptosis in the three clones after transfection of individual PDIA4 or PDIA6 siRNAs and CDDP treatment.
Differential cell death pathways induced by PDIA4 and PDIA6 knockdowns
Further characterization of cellular mechanisms revealed similarities as well as differences in PDIA4 and PDIA6 knockdown-mediated cell deaths. Thus, whereas loss of transmembrane inner potential (ΔΨm; Figure 5a, Supplementary Figures S4A and B) and hypoploidy (Figure 5b) were found in all clones after siRNA PDIA4/CDDP treatment, these events occurred only in R1 following siRNA PDIA6 and CDDP treatment. We then examined the post-mitochondrial caspase activation and found caspases 3/7 activation after siRNA PDIA4 and CDDP treatment and for all resistant clones (Figure 5c, Supplementary Figure S4C and D). However, caspases 3/7 were not activated in any clone by PDIA6 knockdown and CDDP treatment. This result indicates that cell death elicited by siRNA PDIA6/CDDP treatment cannot involve these effector caspases. In contrast, we observed the upregulation of caspase-4, a known inflammation and ER stress-related caspase32, 33 in resistant clones following siRNA PDIA6 and CDPP.
Characterization of cell death elicited by PDIA4 and PDIA6 knockdown and CDDP in resistant cells. (a) Cells transfected with control siRNA (SiCrtl), siRNA targeting PDIA4 (siPDIA4) or siRNA targeting PDIA6 (siPDIA6) for 48 h followed by 48 of 25 μM CDDP were stained with the fluorescent dye TMRM to visualize mitochondrial ΔΨm, (b). with propidium iodide (PI) to visualize DNA content after treatment and analyzed by flow cytometry (n=6, mean=S.D.), and (c). analyzed for caspases 3/7 activity. (d) Western blotting analysis of R1, R2 and R3 cells after 48 h of siRNA PDIA4 or control followed by 48 h of 25 μM CDDP treatment. Bcl-2, caspases 9 and 3 cleavage, GRP78 expression and PARP1 cleavage were analyzed. β-actin is used as a loading control (n=3). (d) Western blotting analysis of R1, R2 and R3 cells after 48 h of siRNA PDIA4 or control followed by 48 h of 25 μM CDDP treatment. GRP78, Bcl-2, Parp1 expression and Caspase 9 and 3 cleavage were analyzed. β-actin is used as a loading control (n=3). (e) Western blotting analysis of R1, R2 and R3 cells after 48 h of siRNA PDIA4 or control followed by 48 h of 25 μM CDDP treatment. Caspase 9, 4 and 3 cleavage, GRP78 expression, PARP1 and RIPK1 expression were analyzed. β-actin is used as a loading control (n=3). (f) Cellular protection by necrostatin of loss of cell viability induced by siRNA PDIA6 and 25 μM CDDP (n=3)
When evaluated by western blotting, PDIA4-dependent cell death pathway recapitulated events that characterize the canonical mitochondrial apoptosis pathway, namely Bcl-2 downregulation, and the cleavage of caspase-3, caspase-9 and PARP1 (Figures 5d and e). In addition, GRP78 expression was slightly enhanced. In contrast, following the treatment siRNA PDIA6/CDDP, no cleavage of the aforementioned proteins was observed, while a marked decrease of GRP78 and an increase in RIPK1 expression was observed (Figure 5e). This is intriguing because RIP1 is a kinase involved in regulated necrosis34 and, accordingly, 50 μM necrostatin 1 blocked the cell death induced in R3 (P<0.01) but not in R2 and R1 (Figure 5f). Moreover, the amount of caspase-4 was upregulated in R2 (3.5 × ) and in R3 (5 × ), along with only a minor increase in the cleaved fragment p10 in R1 cells (Figure 5e).
Pharmacological inhibition of caspases with z-VAD-fmk, a pan-caspase inhibitor, during siPDIA6/CDDP treatment showed that caspases are essential for the cell death process (Figure 6). As caspase-3, -7 and -9 cannot be involved in the lethal pathway (see above), we investigated the role of caspase 4. Here, our results were heterogeneous for the three resistant clones. Z-LEVD-fmk, a specific caspase-4 inhibitor, did not prevent the death after the combined depletion of PDIA6 and CDDP treatment in R1 and R2 clones (Figure 6). In contrast, caspase-4 inhibition significantly inhibited cell death in R3 cells, indicating a specific role of this caspase in this clone. In summary, in contrast to PDIA4 knockdown, which leads to the induction of the mitochondrial cell death pathway, the combination of PDIA6 downregulation and CDDP stimulates cell death through a non-canonical signaling pathway. This shows that that despite apparent similar resistance to CDDP and the major role of the PDI isoforms, the resistance of these clones can affect multiple cell death pathways.
Caspase role in cell death elicited by PDIA4 and PDIA6 knockdown and CDDP. R1, R2 and R3 cells were treated only with siRNA PDIA6 for 48 h followed by 2 h of 50 μM caspase inhibitor Z-VAD or Z-LEVD and/or 48 h of 25 μM CDDP treatment. Cells were labeled with 7-AAD and Annexin V to detect early and late apoptosis (n=6)
Reversal of CDDP resistance in ovarian carcinoma cell line A2780R
To extend our findings concerning the role of PDIA4 and PDIA6 in CDDP resistance, we evaluated the impact of PDIA isoform depletion on A2780R cells, which represent a CDDP-resistant form of an ovarian carcinoma cell line (Supplementary Figure S5A). Signs of CDDP-triggered early and late apoptosis were clearly enhanced by PDI depletion with a stronger impact of PDIA6 than PDIA4 downregulation (76.4 and 44.8%, respectively) (Supplementary Figure S5B). Moreover, an increase of ΔΨm loss and hypoploidy was found following PDI depletion (Supplementary Figures S5C and D), suggesting that PDIA was repressing the mitochondrial apoptotic pathway in A2780R cells.
PDI overexpression in lung adenocarcinoma patients
Finally, the pathophysiological relevance of our study was addressed by immunochemistry in cell lines and lung adenocarcinoma patient biopsies (Supplementary Table S2). In WT and resistant cell lines and patients, we detected a PDIA4-specific cytoplasmic staining pattern compatible with its ER location (Figure 7). PDIA4 was overexpressed in tumoral tissue of CDDP-treated cancer patients (10/10 patients) or CDDP-untreated patients with no mutated EGFR (18/18 patients) but was not detectable in non-tumorous tissue (Figure 7). Unexpectedly, PDIA6 was detected in the nucleus of some patients’ tumor cells, especially in metastatic tumors or those harboring a mutation in EGFR exon 19 or 21 (18/38 lung adenocarcinomas, Supplementary Table S2). Interestingly, 3-fold more untreated CDDP tumors exhibited a nuclear location of PDIA6 than formerly treated and resistant tumors to CCDP. In resistant tumors, PDIA6 was mostly expressed at the cytoplasm of tumoral cells. Moreover, PDIA6 was not detected in the nucleus of normal cells, suggesting tumor-associated nuclear translocation of the protein that could be related to the microenvironment of the tumor.
Immunostaining of patient biopsies and NSLC cell lines. (a) Non-tumor tissue presents substantial negative staining. In this figure, pneumocytes (a) are negative for PDIA4 staining, while a positive staining is seen in macrophages (M) ( × 400). (b) Almost all tumor cells show a PDIA4-positive immunostaining ( × 100). The staining is cytoplasmic with in some cases a membrane reinforcement ( × 400). (c) Non-tumor tissue presents substantial negative staining for PDIA6, with few bronchial cells showing a positive nuclear staining ( × 400). (d and e) Some tumor cells show a PDIA6-positive immunostaining. However, the immunostaining is heterogeneous and varies in different tumor sectors ( × 100), and it is nuclear ( × 400). (f–h) A549 cell lines WT, R1 and R2 show a positive cytoplasmic immunostaining for PDIA4 and PDIA6
Discussion
Chronic treatment of cancer cells with low doses of CDDP prevent apoptosis execution and confer an enhanced survival capacity.5, 16, 22, 35 Here, we found that this resistance phenotype correlate with gene expression regulation of several classical oncogenes and tumor suppressors (e.g., Bax, Bcl-2, PARP1, HSP70) in a panel of resistant NSLC cells. Interestingly, we found a set of overexpressed ER stress proteins, including IRE1, PDIA4 and PDIA6 and downregulated proteins such as GRP78, PERK and ATF6, supporting that ER proteome remodeling, UPR response and PDI proteins might have an original role in cancer cell resistance to apoptosis. Our results demonstrate for the first time that overexpression of two ER proteins, PDIA4 and PDIA6, has a role in acquired resistance to CDDP-induced apoptosis via mechanisms that operate at a pre-mitochondrial level. Despite belonging to the large PDI family, both proteins appear to mediate non-redundant mechanisms of cell death inhibition and to be overexpressed in patient tumor cells. The significance of our findings lies in the clinical importance of chemoresistance for cancer progression and treatment efficacy.
The PDI family is composed of at least 20 proteins involved in the folding and maturation of ER proteins via disulfide formation and cyclic oxidation/reduction.28 Little information is available on the specific role, abundance and regulation of each isoform, but they are described as Ca2+-binding proteins. As a result, it was tempting to hypothesize that their overexpression might lead to a blockade of Ca2+ in the ER and that a defect in Ca2+ fluxes between ER and mitochondria could prevent MMP induction upon CDDP treatment. However, based on ERD1 FRET measurement, no alteration in Ca2+ storage and release by the ER was observed, excluding a link between Ca2+ and CDDP resistance, at least in our cellular model of CDDP resistance.
Evidence for an essential role of PDIA4 and PDIA6 is based on the findings that pharmacological inactivation with bacitracin and genetic silencing with siRNA of these specific isoforms restores the sensitivity to CDDP-induced cell death. This was shown for four NSLC cell lines and one ovarian cancer cell line, establishing the relevance of the observation for various cancer types. Recently, a PDI inhibitor, PACMA 31, has been reported to suppress tumor growth in a mouse xenograft model of human OVCAR-8 ovarian cancer.36 Our results suggest that such a strategy could also be used to reverse CDDP-induced cell death resistance.
Surprisingly, a CDDP-protective function was not observed for other PDIs than PDIA4 and PDIA6. Moreover, mechanisms of PDIA4- and PDIA6-mediated cytoprotection were distinct, notably regarding the role of caspases, ΔΨm dissipation and DNA fragmentation. Although PDIA4 knockdown triggered the classical mitochondrial pathway of apoptosis (ΔΨm loss, proteolytic maturation/activation of caspases 3, 7 and 9, phosphatidylserine exposure and plasma membrane permeabilization and DNA fragmentation), PDIA6 knockdown induced a different mode of cell death with PS exposure and plasma membrane permeabilization, caspase-4 activation and RIPK1 overexpression but no ΔΨm dissipation, caspase-9 cleavage or DNA fragmentation in at least in two out of three clones. This indicates that the specific cytoprotective role of these proteins is not redundant.
Despite a large number of studies linking the UPR to alterations in drug sensitivity and malignancy,9, 10 profound mechanistic insights into these roles have not been established so far. The induction of GRP78 has been commonly used as an indicator for UPR, and the regulation of its expression in resistant cells and its reversal following PDIA4 and PDIA6 depletion suggests that GRP78 may be an important factor in CDDP resistance. Thus, further studies are required that might reveal a major role of GRP78 in chronic chemoresistance. Of note, we also found that the three clones are cross-resistant to other chemotherapeutics such as oxaliplatinum and carboplatinum and that PDIA4 knockdown restored death induction with the same profile than CDDP (Supplementary Figure S6). Only oxaliplatinum was not reversed by PDIA6 extinction in R3 cells.
Importantly, PDIA4 and PDIA6 expression in samples from lung adenocarcinoma patients correlated with their clinical status. We analyzed lung biopsies from patients at diagnosis of non-small cell lung adenocarcinoma or after treatment by CDDP; PDIA4 was found with a cytoplasmic location compatible with the expected ER location in tumor cells from all patients and cell lines. In contrast, PDIA6 can also be detected in the cytoplasm and in the nucleus in some tumor cells in most of the patients. Unusual locations of PDI proteins have been previously proposed to be related either to their redox function or to their ability to bind DNA and nuclear matrix and to activate transcription factors37, 38 or participated to DNA repair. PDI has recently been found in glial cytoplasmic inclusions of patients with multiple system atrophy,39 Alzheimer’s disease40 and amyotrophic lateral sclerosis.41 It is also known that intracellular redistribution of PDIs or modification of its redox activity can be controlled by N-nitrosylation in conditions of nitrosative stress.42, 43, 44 Therefore, it is tempting to speculate that in patient biopsies PDIA6 might redistribute intracellularly as a consequence of yet-to-be elucidated post-translational modifications. The cytosolic location of PDIA6 in CDDP-treated tumors could suggest their role in the DNA damage signaling cascade and provide a novel promising target for chemotherapeutic intervention.45
In summary, we demonstrated overexpression of PDIA4 and PDIA6 in CDDP-resistant NSLC cells and in biopsies from lung adenocarcinoma patients. In vitro, PDIA4- or PDIA6-targeting siRNAs reversed the CDDP-resistant phenotype and established an unexpected anti-apoptotic role of these proteins. We anticipate that this study may prepare the theoretical grounds to define novel chemotherapeutic strategies based on PDI inhibitors.
Materials and Methods
Reagents
Where not indicated, reagents were from Sigma (Saint-Quentin Fallavier, France). Caspase inhibitors Z-VAD-fmk and Z-LEVD-fmk are from BioVision (Lyon, France).
Cell lines
Human A549 and A2780 WT and resistant cells were kindly provided by Professor G Kroemer (Inserm U981, Institut Gustave Roussy, Villejuif, France) and Dr. M Gutmann (Cytomics Pharmaceuticals, Orsay, France). A549 cells were grown in DMEM supplemented with 10% fetal bovine serum, antibiotics and 10% glutamine at 37 °C in a humidified atmosphere with 5% CO2. A2780 cells were maintained in RPMI with 10% fetal bovine serum, antibiotics and 10% glutamine. A549-resistant clones R1, R2 and R3 were obtained as described.16 A2780-resistant cells were maintained with 1 μM CDDP once a week to maintain resistance.
siRNA transfection to knockdown PDI isoforms
On-Targetplus SMART pool siRNA, a mixture of four siRNA provided as a single reagent were purchased from Dharmacon Research (Illkirch, France). The transfections were performed according to Martel et al.46 At day 0, A549 and A2780 cells were plated overnight at 5 × 103 cells in 96-well plates for cell viability and for caspase activity quantification at 3 × 104 in 24-well plates for cytometry analysis. At day 1, the cells were transfected with 100 nM PDI isoforms siRNA oligonucleotides or non-target control oligonucleotides using Dharmafect transfection reagent 2 for 48 h. After siRNA treatment, cells were treated or not with 25 μM CDDP.
Subcellular fractionation
Cells were harvested at 70% confluency in 175 cm3 flasks, trypsinized and washed in PBS. Cells were incubated with 5 ml of buffer (250 mM saccharose, 10 mM Tris pH 7.6, 10 mM KCL, 0.15 mM MgCL2 and 0.4 mM PMSF) for 30 min on ice. Then, cells were broken in Dounce homogenizer (Dominique Dutscher, Brumath, France) (100 hits). Homogenates were centrifuged at 1 000 × g for 10 min to pellet nucleus, cell debris and intact cells. The supernatants were collected and centrifuged twice at 10 000 (mitochondria fraction) and 20 000 × g (mitochondria and membranes debris fraction) for 15 min at 4 °C to pellet mitochondria. Again, the supernatants were centrifuged at 100 000 × g at 4 °C to pellet ER. Finally, cytosolic proteins were precipitated with acetone. ER was solubilized overnight in proteomic buffer (7 M urea, 2 M thio-urea, 20 mM dithiothreitol (DTT), 1%Triton X-100, ampholytes 3–10; Bio-Rad, Marnes la Coquette, France) and centrifuged for 45 min at 100 000 × g at 4 °C.
Proteomic sample preparation
ER proteins (125 μg) were diluted in proteomic buffer supplemented with bromophenol blue in a final volume of 150 μl and loaded overnight on a 17 cm Ready IPG strip 2D electrophoresis pH3-10 (Bio-Rad) for rehydratation in a passive mode. The rehydrated strip with protein was placed in Protean IEF cell (Bio-Rad), and proteins were separated following iso-focalization. Strips were equilibrated 10 min in 5 ml of buffer containing 50 mM Tris-HCl, pH8.8, 6 M urea, 2% SDS, 30% glycerol and 1% DTT. A second equilibration with bromophenol blue was performed in 5 ml of the same buffer with 1.5% iodoacetamide substituted for DTT during 10 min. Strips were transferred onto a 12%-SDS-polyacrylamide gel and overlay with 0.5% agarose in Laemmli buffer. Second-dimension denaturing electrophoresis was driven at 200 V, and gels were stained with silver nitrate in small Dodeca stainer (Bio-Rad). Gels were scanned with GS800 Calibrated densitometer (Bio-Rad), and candidate spots are analyzed with PDQuest software (Bio-Rad).
Mass spectrometry analysis
Spots of interest were in-gel digested with sequence-grade trypsin (Promega, Madison WI, USA) and analyzed by tandem mass spectrometry (MS) as previously described.46 The peptide mass profiles obtained were analyzed using MASCOT MS/MS Ion Search (http://www.matrixscience.com) with the following parameters: Swiss-Prot 57.7 database, Homo sapiens taxonomy, one missed cleavage by trypsin, fixed carbamidomethylation of cysteine and variable oxidation of methionine, monoisotopic peptide masses, peptide tolerance of 100 p.p.m. and MS/MS tolerance of 0.5 Da. Validated proteins had at least one peptide matched and a minimum Mascot score of 32 (significance threshold P<0.05).
Western blotting analysis
Cells (300 000) were collected and lyzed with RIPA buffer. An equal amount of proteins (30 μg) was separated by SDS-PAGE and immunoblotted with the following antibodies: ATF6 (Abcam, Thermo Fischer, Illkirch, France); Bax, (BD Biosciences, Heidelberg, Germany); Bcl-2, (Santa Cruz, Saint Quentin, France); Caspase 9, (Abcam); GRP78 (Cell Signaling, Paris, France); HSP70 (Santa Cruz); IRE1 (Abcam); PARP1 (Santa Cruz); PDIA1 (Assay Design Ann Arbor, MI, USA); PDIA3 (Abcam); PDIA4 (Abcam and Santa Cruz); PDIA6 (Santa Cruz and Abcam); PERK (Abcam); and RIPK1 (BioVision).
Cell viability analysis
The number of surviving cells was determined by the WST-1 assay (Roche, Boulogne-Bilancourt, France).
Cell death analysis
A549 and A2780 WT and resistant cells were analyzed for cell death as previously described.47 Briefly, tetramethylrhodamine methyl ester (TMRM, Invitrogen, Saint Aubin, France) was used to measure the mitochondrial transmembrane potential (ΔΨm) at 24 h, propidium iodide for cell cycle and subG1 (hypoploidy) at 48 h by flow cytometry (FACSCalibur flow cytometer, BD Biosciences). Finally, early and late apoptosis were analyzed using Annexin-PE/7AAD (BD Pharmingen, San Jose, CA, USA). The caspases 3/7 activities were measured with the Apo-ONE Homogeneous Caspase-3/7 Assay (Promega, San Jose, CA, USA).
PDI activity assay
ProteoStat PDI assay kit was used following the manufacturer’s instruction (Enzo Life, Villeurbanne, France) by using insulin as substrate to evaluate PDI activity in A549 whole extracts and ER fraction.
Clonogenic assay
Cells were harvested, washed with PBS and plated for 2–3 weeks at 37 °C. Then, colonies were stained with 0.25% of crystal violet and counted using GS800 calibrated densitometer (Bio-Rad) and PDQuest software (Bio-Rad).
[Ca2+] measurements in the ER and cytoplasm
For steady state [Ca2+] ER measurements, the ERD1 FRET-based recombinant Ca2+ probe was used, as previously described.26 Briefly, cells were plated on 22 mm glass coverslips and, 48 h following transfection, were treated or not with 75 μM CDDP for 24 h. Measurements were carried out on a Zeiss LSM510 META confocal system (Marly le Roy, France). The probe was excited by a 405-nm laser diode, emission spectra were acquired from 420–600 nm and the YFP and CFP signals were obtained by un-mixing the spectrum based on previously registered spectra of separate CFP and YFP proteins, as well as the autofluorescence of non-transfected cells. FRET efficiency, which is the function of the ER luminal [Ca2+] was quantified using the acceptor bleaching method.48 Briefly, after five acquisitions YFP was bleached (at both 488 and 514 nm excitation wavelengths, typically by about 80–90%), followed by acquisition of further five image spectra. Reduction of the YFP signal leads to an increase in the CFP signal, which was normalized to the decrease of YFP intensity during bleaching. The normalized increase of CFP intensity is presented as FRET efficiency (Supplementary Figure S1).
For cytosolic [Ca2+] measurements, A549-WT and R1 cells were seeded in 24-well plates. Cells were loaded with 1 μM Fura-2-AM (Invitrogen) at 37 °C for 20 min. Cells were imaged on Olympus IX71 inverted epifluorescence microscope (Olympus, Hamburg, Germany) fitted with a computer-controlled motorized heated stage (37 °C, Applied Scientific Instrumentation, Eugene, OR, USA), with a × 40 fluorite objective lens (NA 0.6). Images were collected using a Hamamatsu C10600-10B CCD camera (Hamamatsu Corporation, Massy, France), and recorded and analyzed using the Simple PCI 6.6.0.0 software (Hamamatsu Corporation). Excitation illumination was provided by light from a metal halide arc lamp passing through a computer-controlled filter wheel (Prior Scientific, Rockland, MD, USA), using a Fura-2 filter set (71 000, Chroma Technologies, Bellows Falls, VT, USA). Following 10 min of image acquisition to determine baseline Fura-2 fluorescence, 75 μM CDDP was added directly to the appropriate samples, and data were acquired for a further 60 min. Data were acquired at the same time point for each test condition from cells in adjacent wells of the tissue culture plate loaded simultaneously with Fura-2-AM. Statistical significance was assessed through comparison of the background-corrected 340 nm: 380 nm Fura-2 fluorescence ratio of the final 10 frames of the image series using a paired Student’s t-test.
Immunohistology
Forty-two patient lung biopsies from Hospital P Brousse, Villejuif, France and cells from WT and R1 cell lines were analyzed. Immunohistology was performed on 4-μm paraffin-embedded formalin-fixed biopsies or cell pellet sections using anti-PDIA4 (Abcam 82587) and PDIA6 (Abcam 89668) antibodies. Antigen retrieval was obtained by heat at 97 °C in a citrate buffer at pH 6. The revelation system was based on a one-step biotin-free immunoperoxydase stain (Envision, DAKO, Glostrup, Denmark) using 3,3-diamino-benzidine chromogene (DAKO, Glostrup, Denmark) substrate followed by Hemalun counterstaining. Negative controls for each slide were processed concurrently with probed samples by omitting primary antibody.
Statistical analysis
Data were analyzed using Student’s t-test for all pair-wise comparisons of growth rates, and mean responses to the different treatments were tested. Results are presented as the mean±S.D. of three replicate experiments. Significance: *P<0.05; **P<0.01; ***P<0.001.
Abbreviations
- ATF6:
-
activating transcription factor 6
- CDDP:
-
cis-diaminedichloroplatine(II)
- DTT:
-
dithiothreitol
- IRE1:
-
ER to nucleus signaling 1
- MMP:
-
mitochondrial membrane permeabilization
- MS:
-
mass spectrometry
- NSLC:
-
non-small lung cancer
- PDI:
-
protein disulfide isomerase
- PERK:
-
protein kinase RNA-like endoplasmic reticulum kinase
- GRP78:
-
78 kDa glucose-regulated protein
- ΔΨm:
-
transmembrane inner potential
References
Kroemer G, Galluzzi L, Brenner C . Mitochondrial membrane permeabilization in cell death. Physiol Rev 2007; 87: 99–163.
Hanahan D, Weinberg RA . The hallmarks of cancer. Cell 2000; 100: 57–70.
Rebillard A, Tekpli X, Meurette O, Sergent O, LeMoigne-Muller G, Vernhet L et al. Cisplatin-induced apoptosis involves membrane fluidification via inhibition of NHE1 in human colon cancer cells. Cancer Res 2007; 67: 7865–7874.
Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O et al. Molecular mechanisms of cisplatin resistance. Oncogene 2012; 31: 1869–1883.
Köberle B, Tomicic M, Usanova S, Kaina B . Cisplatin resistance: preclinical findings and clinical implications. Biochim Biophys Acta 2010; 1806: 172–182.
Ma Y, Hendershot LM . The role of the unfolded protein response in tumour development: friend or foe? Nat Rev 2004; 4: 966–977.
Zhang K, Kaufman RJ . Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem 2004; 279: 25935–25938.
Ron D, Walter P . Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007; 8: 519–529.
Hersey P, Zhang XD . Adaptation to ER stress as a driver of malignancy and resistance to therapy in human melanoma. Pigment Cell Melanoma Res 2008; 21: 358–367.
Kim I, Xu W, Reed J . Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov 2008; 7: 1013–1030.
Deniaud A, Sharaf el dein O, Maillier E, Poncet D, Kroemer G, Lemaire C et al. Endoplasmic reticulum stress induces calcium-dependent permeability transition, mitochondrial outer membrane permeabilization and apoptosis. Oncogene 2008; 27: 285–299.
Lee AS . GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res 2007; 67: 3496–3499.
Han W, Xie J, Li L, Liu Z, Hu X . Necrostatin-1 reverts shikonin-induced necroptosis to apoptosis. Apoptosis 2009; 14: 674–686.
Kim Y, Haidl G, Schaefer M, Egner U, Herr J . Compartmentalization of a unique ADP/ATP carrier protein SFEC (Sperm Flagellar Energy Carrier, AAC4) with glycolytic enzymes in the fibrous sheath of the human sperm flagellar principal piece. Dev Biol 2007; 302: 463–476.
Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM et al. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol 2006; 4: e374.
Yao Z, Jones A, Fassone E, Sweeney M, Lebiedzinska M, Suski JM et al. PGC-1β mediates adaptive chemoresistance associated with mitochondrial DNA mutations. Oncogene 2013; 32: 2592–2600.
Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 2012; 19: 107–120.
Tajeddine N, Galluzzi L, Kepp O, Hangen E, Morselli E, Senovilla L et al. Hierarchical involvement of Bak, VDAC1 and Bax in cisplatin-induced cell death. Oncogene 2008; 27: 4221–4232.
Belfi CA, Chatterjee S, Gosky DM, Berger SJ, Berger NA . Increased sensitivity of human colon cancer cells to DNA cross-linking agents after GRP78 up-regulation. Biochem Biophys Res Commun 1999; 257: 361–368.
Konopleva M, Zhao S, Hu W, Jiang S, Snell V, Weidner D et al. The anti-apoptotic genes Bcl-X(L) and Bcl-2 are over-expressed and contribute to chemoresistance of non-proliferating leukaemic CD34+ cells. Br J Haematol 2002; 118: 521–534.
Ravagnan L, Gurbuxani S, Susin SA, Maisse C, Daugas E, Zamzami N et al. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat Cell Biol 2001; 3: 839–843.
Michels J, Vitale I, Senovilla L, Enot D, Garcia P, Lissa D et al. Synergistic interaction between cisplatin and PARP inhibitors in non-small cell lung cancer. Cell Cycle 2013; 12: 877–883.
Mandic A, Hansson J, Linder S, Shoshan MC . Cisplatin induces endoplasmic reticulum stress and nucleus-independent apoptotic signaling. J Biol Chem 2003; 278: 9100–9106.
Chami M, Oulès B, Szabadkai G, Tacine R, Rizzuto R, Paterlini-Bréchot P . Role of SERCA1 truncated isoform in the proapoptotic calcium transfer from ER to mitochondria during ER stress. Mol Cell 2008; 32: 641–651.
Palmer AE, Jin C, Reed JC, Tsien RY . Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc Natl Acad Sci USA 2004; 101: 17404–17409.
Vicencio J, Ortiz C, Criollo A, Jones A, Kepp O, Galluzzi L et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ 2009; 16: 1006–1017.
Inesi G, Sagara Y . Thapsigargin, a high affinity and global inhibitor of intracellular Ca2+ transport ATPases. Arch Biochem Biophys 1992; 298: 313–317.
Schwaller M, Wilkinson B, Gilbert HF . Reduction-reoxidation cycles contribute to catalysis of disulfide isomerization by protein-disulfide isomerase. J Biol Chem 2003; 278: 7154–7159.
Roth RA . Bacitracin: an inhibitor of the insulin degrading activity of glutathione-insulin transhydrogenase. Biochem Biophys Res Commun 1981; 98: 431–438.
Lovat P, Corazzari M, Armstrong J, Martin S, Pagliarini V, Hill D et al. Increasing melanoma cell death using inhibitors of protein disulfide isomerases to abrogate survival responses to endoplasmic reticulum stress. Cancer Res 2008; 68: 5363–5369.
Ellgaard L, Frickel EM . Calnexin, calreticulin, and ERp57: teammates in glycoprotein folding. Cell Biochem Biophys 2003; 39: 223–247.
Bian ZM, Elner SG, Elner VM . Dual involvement of caspase-4 in inflammatory and ER stress-induced apoptotic responses in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci 2009; 50: 6006–6014.
Hitomi J, Katayama T, Eguchi Y, Kudo T, Taniguchi M, Koyama Y et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J Cell Biol 2004; 165: 347–356.
Vandenabeele P, Declercq W, Van Herreweghe F, Vanden Berghe T . The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci Signal 2010; 3: re4.
Michels J, Vitale I, Galluzzi L, Adam J, Olaussen K, Kepp O et al. Cisplatin resistance associated with PARP hyperactivation. Cancer Res 2013; 73: 2271–2280.
Xu S, Butkevich A, Yamada R, Zhou Y, Debnath B, Duncan R et al. Discovery of an orally active small-molecule irreversible inhibitor of protein disulfide isomerase for ovarian cancer treatment. Proc Natl Acad Sci USA 2012; 109: 16348–16353.
Turano C, Coppari S, Altieri F, Ferraro A . Proteins of the PDI family: unpredicted non-ER locations and functions. J Cell Physiol 2002; 193: 154–163.
VanderWaal R, Spitz D, Griffith C, Higashikubo R, Roti Roti J . Evidence that protein disulfide isomerase (PDI) is involved in DNA–nuclear matrix anchoring. J Cell Biochem 2002; 85: 689–702.
Honjo Y, Ito H, Horibe T, Takahashi R, Kawakami K . Protein disulfide isomerase immunopositive glial cytoplasmic inclusions in patients with multiple system atrophy. Int J Neurosci 2011; 121: 543–550.
Honjo Y, Ito H, Horibe T, Takahashi R, Kawakami K . Protein disulfide isomerase-immunopositive inclusions in patients with Alzheimer disease. Brain Res 2010; 1349: 90–96.
Walker A, Farg M, Bye C, McLean C, Horne M, Atkin J . Protein disulphide isomerase protects against protein aggregation and is S-nitrosylated in amyotrophic lateral sclerosis. Brain Res 2010; 133: 105–116.
Uys J, Xiong Y, Townsend D . Nitrosative stress-induced S-glutathionylation of protein disulfide isomerase. Methods Enzymol 2011; 490: 321–332.
Bernardoni P, Fazi B, Costanzi A, Nardacci R, Montagna C, Filomeni G et al. Reticulon1-C modulates protein disulphide isomerase function. Cell Death Dis 2013; 4: e581.
Nakamura T, Lipton S . Redox modulation by S-nitrosylation contributes to protein misfolding, mitochondrial dynamics, and neuronal synaptic damage in neurodegenerative diseases. Cell Death Differ 2011; 18: 1478–1486.
Krynetskaia N, Phadke M, Adhav S, Krynetskiy E . Chromatin-associated proteins HMGB1/2 and PDIA3 trigger cellular response to chemotherapy-induced DNA damage. Mol Cancer Ther 2009; 4: 864–872.
Martel C, Allouche M, Esposti DD, Fanelli E, Boursier C, Henry C et al. GSK3-mediated VDAC phosphorylation controls outer mitochondrial membrane permeability during lipid accumulation. Hepatology 2013; 57: 93–102.
Le Bras M, Borgne-Sanchez A, Touat Z, Sharaf el dein O, Deniaud A, Maillier E et al. Chemosensitization by knock-down of adenine nucleotide translocase-2. Cancer Res 2006; 66: 9143–9152.
Gu Y, Di W, Kelsell D, Zicha D . Quantitative fluorescence resonance energy transfer (FRET) measurement with acceptor photobleaching and spectral unmixing. J Microsc 2004; 215: 162–173.
Acknowledgements
We thank Dr. F Poirier for her technical assistance and Dr. L Galluzzi for the generous gift of resistant cell lines. We are supported by the European Commission (ArtForce); European Research Council, Agence National de la Recherche (ANR); Ligue contre le Cancer (Equipe labellisée); Fondation pour la Recherche Médicale (FRM); Institut National du Cancer (INCa); LabEx LERMIT; LabEx Immuno-Oncologie; Fondation de France; Fondation Bettencourt-Schueller; AXA Chair for Longevity Research; and Cancéropôle Ile-de-France and Paris Alliance of Cancer Research Institutes (PACRI). GS is supported by Parkinson’s UK, Wellcome Trust, Italian Association of Cancer Research (AIRC) and Telethon Italy. ZW receives a fellowship from the China Government.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by P Mehlen
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Tufo, G., Jones, A., Wang, Z. et al. The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma. Cell Death Differ 21, 685–695 (2014). https://doi.org/10.1038/cdd.2013.193
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2013.193
Keywords
This article is cited by
-
Protein disulfide isomerase family member 4 promotes triple-negative breast cancer tumorigenesis and radiotherapy resistance through JNK pathway
Breast Cancer Research (2024)
-
Protein disulfide-isomerase A4 confers glioblastoma angiogenesis promotion capacity and resistance to anti-angiogenic therapy
Journal of Experimental & Clinical Cancer Research (2023)
-
PDIA4 confers resistance to ferroptosis via induction of ATF4/SLC7A11 in renal cell carcinoma
Cell Death & Disease (2023)
-
Deep topographic proteomics of a human brain tumour
Nature Communications (2023)
-
Protein disulfide isomerase blocks the interaction of LC3II-PHB2 and promotes mTOR signaling to regulate autophagy and radio/chemo-sensitivity
Cell Death & Disease (2022)