Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and its receptors, TRAIL-R1 (DR4) and TRAIL-R2 (DR5), promote the selective clearing of various malignancies by inducing apoptosis, holding the promise as a potent therapeutic agent for anticancer. Though DR4 and DR5 have high sequence similarity, differential regulation of both receptors in human tumor cells remains largely unexplored. Here, we repot that golgi-specific Asp-His-His-Cys (DHHC) zinc finger protein (GODZ) regulates TRAIL/DR4-mediated apoptosis. Using the SOS protein recruitment–yeast two-hybrid screening, we isolated GODZ that interacted with the death domain of DR4. GODZ binds to DR4, but not to DR5, through the DHHC and the C-terminal transmembrane domain. Expression level of GODZ affects apoptosis of tumor cells triggered by TRAIL, but not that induced by TNF-α/cycloheximide (CHX) or DNA-damaging drugs. In parallel, GODZ functions to localize DR4 to the plasma membrane (PM) via DHHC motif. Also, introduction of mutation into the cysteine-rich motif of DR4 results in its mistargeting and attenuates TRAIL- or GODZ-mediated apoptosis. Interestingly, GODZ expression is highly downregulated in Hep-3B tumor cells, which show resistance to TRAIL. However, reconstitution of GODZ expression enhances the targeting of DR4 to cell surface and sensitizes Hep-3B cells to TRAIL. Taken together, these data establish that GODZ is a novel DR4-selective regulator responsible for targeting of DR4 to the PM, and thereby for TRAIL-induced apoptosis.
Similar content being viewed by others
Main
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), a member of the TNF superfamily, has strong antitumor activity in a wide variety of cancer cells and minimal cytotoxicity in most normal cells. TRAIL and agonistic antibodies against its receptor are thus currently being studied in clinical trials as a cancer treatment alone or in combination with various chemotherapeutic agents. TRAIL is a ligand that initiates extrinsic apoptosis by binding to two cognate receptors, TRAIL-R1 (DR4) and TRAIL-R2 (DR5), in human.1, 2 TRAIL can signal both apoptosis in transformed tumor cells and non-apoptotic signaling cascades. Clustering and oligomerization of DR4 and DR5 by TRAIL result in recruitment of various adaptor proteins, including FADD and proximal caspases to form a death-inducing signaling complex (DISC), leading to the activation of caspases-8 and -10 within DISC.3, 4, 5 In addition, TRAIL receptors also recruit TRADD and RIP kinase to signal NF-κB activation through its death domain.6, 7 The mouse genome has a single TRAIL receptor resembled with DR5 sequences,8 and its deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis.9
Cancer cells have acquired resistance to cell death by inducing varied strategies.10 Signaling to cell death can be inhibited by an increase of antiapoptotic protein or by a decrease of or defective proapoptotic proteins. It has been reported that simultaneous expression of antagonistic decoy receptors DcR1 (TRAIL-R3) and DcR2 (TRAIL-R4), mutational inactivation of DR4 and DR5, dysfunction of DISC components (for example, FLIP, FADD, caspases-8 and -10), and overexpression of Bcl-2 family proteins are attributed to TRAIL resistance in cancer cells.3, 5, 11 Both DR4 and DR5 have also been demonstrated to be induced by many chemotherapeutic drugs, ER stress inducer, and IFN-γ12, 13 and transcriptionally regulated by p53 and NF-κB.14, 15 Thus, studies on TRAIL as a chemotherapeutic drug have focused on combination therapy for regulating proapoptotic and antiapoptotic activity to synergistically increase the tumor sensitivity to TRAIL.16, 17, 18 In addition, post-translational modification of DR4 and DR5 by GALNT14 (O-glycosyltransferase) promotes ligand-stimulated clustering of those death receptors and TRAIL-sensitivity in tumor cells.19 We also recently reported that peroxiredoxin 6 and ribosomal protein S6 regulate TRAIL sensitivity of tumor cells.20, 21 However, molecular detail for the resistance of tumor cells to TRAIL is not known completely yet, and overcoming of the resistance will be essential to understand tumorigenesis and for therapeutic approaches in different types of cancer cells.
There are increasing numbers of Asp-His-His-Cys (DHHC) motif-containing proteins. Until now, 24 distinct mammalian DHHC proteins have been identified. DHHC proteins are implicated for their roles in membrane targeting or trafficking of diverse substrates, and a few of them are associated with tumorigenic activity. For example, DHHC-2 facilitates trafficking of CKAP4 from the ER to the plasma membrane (PM) by palmitoylation and is believed to function as a tumor suppressor in colorectal cancers.22, 23 DHHC-17 (HIP 14) that is upregulated in various cancer types affects the PM localization of RAS protein and may thus act as an oncogene.24 Golgi apparatus-specific protein with a DHHC zinc finger domain (GODZ, also known as DHHC-3) was reported as a palmitoyl acyltransferase (PAT) that palmitoylates the cytoplasmic loop domain of the substrates.25 GODZ has a cysteine-rich domain (CRD) with DHHC motif, which has a PAT activity for targeting NMDA receptor.26, 27, 28 However, the role of DHHC protein(s) in TRAIL-mediated apoptosis is not known.
To better understand TRAIL-mediated tumor cell death, we have searched for membrane-associated protein that interacted with the DR4. We isolated GODZ as a novel modulator in the TRAIL/DR4 signaling. GODZ regulates PM targeting of DR4 and thus TRAIL/DR4-mediated apoptosis in cancer cells.
Results
GODZ binds to DR4 via CRD domain-containing C-terminus, but not to DR5
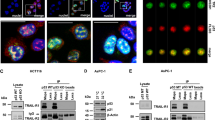
To excavate novel membrane-associated protein(s) that interacts with DR4, we undertook the SOS protein recruitment–yeast two-hybrid screening, which was designed to isolate the membrane protein. Using the death domain of DR4 as bait, we isolated GODZ from a human thymus cDNA library as a DR4-interacting protein in yeast (Supplementary Figure S1a). GODZ is an integral membrane protein with four putative transmembrane domains and a DHHC-containing CRD at the middle region. At first, the interaction of HA-tagged GODZ with Flag-tagged DR4 was determined in mammalian cells by the immunoprecipitation assays using anti-Flag antibody. From western blot analysis, we found GODZ-HA in the immunoprecipitates containing DR4-Flag (Figure 1a). In the reverse immunoprecipitation assays, we also found DR4-Flag in the GODZ protein complex (Figure 1b). However, we could not detect DR5-Flag in the GODZ-containing immunoprecipitates. Moreover, when we analyzed the formation of endogenous protein complexes using the same assays with an antibody against either GODZ or DR4, we consistently found that GODZ interacted with DR4, but not with DR5, in HeLa cells (Figure 1c). These results suggest that GODZ interacts selectively with DR4 in cells.
GODZ binds to DR4 but not to DR5 in cells. (a and b) Reciprocal immunoprecipitation assays using HA- or Flag-tagged proteins. HEK 293T cells were transfected with pGODZ-HA, pDR4-Flag, or empty vector for 24 h and cell extracts were subjected to immunoprecipitation (IP) with anti-Flag (M2) antibody (a) or monoclonal anti-HA antibody (b) The immunoprecipitates were analyzed by western blotting using the indicated antibodies. Whole cell lysates (WCL) are indicated. Heavy chain (*) and light chain (**) of immnoglobulins were served as loading control. The arrowhead in (b) indicates nonspecific signal of the immunoprecipitates. (c) Cellular interaction between endogenous GODZ and DR4. HeLa cells extracts were prepared and IP with pre-immune serum (Pre) or anti-GODZ antibody (upper) and Pre or anti-DR4 antibody (lower). The immunoprecipitates were then analyzed by western blotting using anti-GODZ, anti-DR4, or anti-DR5 antibody. The asterisk indicates light chain (*) of immunoglobulins. (d) Visualization of the association of GODZ with DR4 in living cells using BiFC assay. HeLa cells were transiently co-transfected with equimolar amounts of pGODZ-VN and pDR4-VC, pGODZ-VN and pDR5-VC, pbJun-VN and pbFos-VC (a positive control), or pVN and pVC (a negative control). Green fluorescence images of the complementation were acquired under a fluorescence microscope (GFP; left) and then overlapped with Hoechst 33258 staining for nuclei (Hoechst, middle; and Overlay, right). The color reproduction of this figure is available at the Cell Death and Differentiation journal online
Further, we investigated the direct interaction between DR4 and GODZ in living cells using a split-Venus-based bimolecular fluorescence complementation (BiFC) assay,29 which enabled us to visualize formation of the protein complexes and localization of the protein interaction in living cells by fluorescence resonance energy transfer. Each of the TRAIL receptors was fused with the C-terminal half of Venus vector (DR4-VC and DR5-VC). These fusion cDNAs were transfected into HeLa cells expressing GODZ fused with the N-terminal half of Venus (GODZ-VN). Coexpression of DR4-VC and GODZ-VN exhibited fluorescence complementation, which was observed as scattered green fluorescence dominantly in the Golgi apparatus and PM, whereas coexpression of DR5-VC with GODZ-VN showed very weak fluorescence (Figure 1d). Expressions of these cDNAs were confirmed by western blotting (data not shown). The fluorescence complementation between the bZIP domains of Jun and Fos (bJun and bFos) fused to pVN and pVC (bJun-VN and bFos-VC, respectively) was used as a positive control and observed in nucleoli forming the green dots. In addition, immunocytochemical analysis showed subcellular localization of GODZ-HA in the Golgi apparatus where DR4-GFP also colocalized (Supplementary Figures S1b and c).
In order to map the domains of GODZ, which interact with DR4, we generated HA-tagged deletion mutants of GODZ on the basis of predicted secondary structure such as transmembrane and the CRD (Figure 2a, left) and confirmed their expressions in HEK 293T cells by western blotting (Figure 2b, left). From the immunoprecipitation assays using anti-HA antibody, we found that GODZ-A mutant also interacted with DR4, showing that deletion of the N-terminus (amino-acid residues 1–95) did not affect the ability of GODZ to interact with DR4 (Figure 2b, right). When the CRD was further deleted from GODZ-A mutant (GODZ-C) or the C-terminus (amino-acid residues 170–299) of GODZ was deleted (GODZ-B), however, the capacities of these mutants to interact with DR4 were lost. In the reverse immunoprecipitation assays using anti-Flag (M2) antibody, we found similar results; GODZ-A mutant bound to DR4 (Figure 2c). These data suggest that the DHHC domain and the C-terminal transmembrane domain of GODZ are required for the binding to DR4.
CRD domain-containing C-terminus of GODZ is necessary for the interaction with DR4. (a) Schematic diagram of GODZ functional domains. Numbers indicate amino-acid residues of GODZ. (b and c) Mapping of GODZ domains for the interaction with DR4. HEK 293T cells were transfected with pDR4-Flag (DR4-Flag) alone or together with either pGODZ-HA or pGODZ deletion mutant for 24 h, and cell extracts were IP with anti-HA antibody (b) or anti-Flag (M2) antibody (c). The immunoprecipitates were then analyzed by western blotting using anti-Flag or anti-HA antibody. WCL are indicated and heavy chains (*) of immunoglobulins are served as loading control. TM, transmembrane domain; CRD, cysteine-rich domain
Expression of GODZ regulates TRAIL-mediated cell death via DR4
We then addressed the role of GODZ in TRAIL-induced cell death. Using GODZ shRNAs #1 and #2, we generated HeLa-stable cells (HeLa/GODZ shRNA #1 and #2 cells, respectively), which show the reduced expression of GODZ (Figure 3a). Knockdown of GODZ expression itself did not affect cell’s viability in the absence of cell death signal. Also, we confirmed that GODZ did not disturb mRNA level of DR4. Interestingly, compared with control cells, HeLa/GODZ shRNA #1 and #2 cells became significantly resistant to cell death triggered by TRAIL (Figure 3b). On the other hand, there was no difference in cell's sensitivity to the extrinsic apoptosis triggered by TNF-α with CHX or the intrinsic apoptosis by etoposide (Etopo), tunicamycin (Tuni), or doxorubicin (Doxo) (Figure 3c). After incubation with TRAIL, we confirmed that the proteolytic activation of caspases-8 and -10 and PARP-1 cleavage, a marker of caspase-3 activation, were apparently reduced in HeLa/GODZ knockdown cells, as examined with western blot analysis (Figure 3d). These data suggest that GODZ has an essential role in TRAIL-mediated apoptosis.
Down-regulation of GODZ expression desensitizes tumor cells to apoptosis induced by TRAIL but not by other signals. (a) Generation of HeLa/GODZ knockdown cells. HeLa cells were transfected with control vector (pCtrl shRNA) or two different kinds of pGODZ shRNAs for 24 h and then selected with G418 (1 mg/ml) for 2 weeks to generate mixed cell population (HeLa/Ctrl shRNA, HeLa/GODZ shRNA #1 or #2), after which cell extracts were prepared and the levels of proteins were examined by western blotting with anti-GODZ and anti-β-actin antibodies (upper, 2 panels). Total RNA was prepared and examined for the mRNA levels of GODZ and DR4 by RT-PCR analysis using gene-specific primers (lower, 3 panels). (b) Knockdown of GODZ expression desensitizes HeLa cells to TRAIL. HeLa/Ctrl shRNA control cells and HeLa/GODZ shRNA cells (#1 and #2) were treated with TRAIL (100 ng/ml) for the indicated times. After staining with EtHD, cell death was quantified by counting the number of EtHD-positive cells among total cells. Values indicate mean±S.D. (n=3). P values were estimated using t-test versus control. (c) Selective resistance of HeLa/GODZ knockdown cells to TRAIL. HeLa cells were co-transfected with pEGFP (Clontech) and either pCtrl shRNA or pGODZ shRNAs for 48 h, and then left untreated (MOCK) or treated with TRAIL (100 ng/ml) for 3 h, TNF-α (30 ng/ml) with cycloheximide (CHX) (5 μg/ml) for 3 h, etoposide (25 μM, etopo.) for 24 h, tunicamycin (2 μg/ml, tuni.) for 24 h, or doxorubicin (125 ng/ml, doxo.) for 24 h. Cell death rates were quantified as described in (b). Bars indicate mean±S.D. (n=3). P values were estimated using t-test versus control (*P<0.05; **P<0.01). (d) Suppression of caspase-8 activation in HeLa/GODZ knockdown cells by TRAIL. HeLa/Ctrl shRNA control cells and HeLa/GODZ shRNA #1 cells were left untreated or incubated with TRAIL (100 ng/ml) for 3 h, and cell extracts were examined by western blotting using the anti-PARP-1, anti-caspase-8, anti-caspase-10, anti-DR4, and anti-β-actin antibodies
Conversely, when we ectopically expressed GODZ, HeLa cells were healthy without any treatment but became significantly sensitive to cell death triggered by TRAIL (Figure 4a). Compared with control cells, enzymatic activation of caspases-3 and -8 was observed at early times in these cells showing GODZ overexpression (Figure 4c). On the other hand, the increased expression of GODZ did not affect cell death induced by other signals, including TNF-α with CHX, Etopo, Tuni, or Doxo (Figure 4b), consistent to the results observed in GODZ knockdown cells (Figure 3c). On the other hand, GODZ-A or GODZ-B deletion mutant lacking either the C- or N-terminus was not able to sensitize TRAIL-induced apoptosis (Figure 4d), though GODZ-A mutant bound to DR4. Because most of tumor cells express both TRAIL receptor DR4 and DR5, the differential effects of GODZ on TRAIL receptors were further addressed in DR4 or DR5 knockdown cells. Downregulation of DR5 expression using DR5-targeted shRNA suppressed TRAIL-induced apoptosis, but significantly potentiated the cell death after overexpression of GODZ in HeLa cells (Figure 4e). On the contrary, TRAIL-induced apoptosis was not much potentiated by GODZ overexpression in HeLa/DR4 knockdown cells (Figure 4f). Slight increase of TRAIL-cell death by GODZ overexpression in HeLa/DR4 knockdown cells might result from incomplete knockdown of DR4 expression in HeLa cells. We detected the similar expression level of GODZ protein in DR5 and DR4 knockdown cells (Supplementary Figures S2a and b). On the basis of our combined data, we propose that GODZ potentiates exclusively TRAIL-induced apoptosis through DR4.
Increased expression of GODZ sensitizes tumor cells to TRAIL or DR4. (a) Ectopic expression of GODZ increases TRAIL-mediated apoptosis. HeLa cells were transiently co-transfected with pEGFP (Clontech) and either pcDNA3-HA (Ctrl) or pGODZ-HA for 24 h, and then left untreated (MOCK) or treated with TRAIL (100 ng/ml) for the indicated times. Cell death rates were measured by counting the number of both GFP- and EtHD-positive cells among total GFP-positive cells after staining with EtHD. Values indicate mean±S.D. (n=3). (b) Overexpression of GODZ sensitizes HeLa cells to TRAIL, but not to other insults. HeLa cells were co-transfected with pEGFP (Clontech) and either pcDNA3-HA (Ctrl) or pGODZ-HA for 24 h, and then treated with TRAIL (100 ng/ml) or TNF-α (30 ng/ml) with CHX (5 μg/ml) for 3 h, 25 μM etopo or 2 μg/ml tuni. for 24 h, or 125 ng/ml doxo for 24 h. Cell death ratios were measured as described in (a). Bars indicate mean±S.D. (n=3). P values were estimated using t-test versus control. (c) Enhanced activation of caspases by GODZ. HeLa cells transiently transfected with pcDNA3-HA (Ctrl) or pGODZ-HA for 24 h were treated with TRAIL (100 ng/ml) for the indicated times. Western blotting was performed with the indicated antibodies. (d) Effects of GODZ deletions on TRAIL sensitivity. HeLa cells were transiently co-transfected with pEGFP (Clontech) and one of GODZ deletion mutants for 24 h, and then exposed to TRAIL (100 ng/ml) for additional 3 h. Cell death ratios were measured as described in (b). Bars indicate mean±S.D. (n=3). P-values were estimated using t-test versus control (*P<0.05; **P<0.01). (e and f) Distinct sensitivity of DR4 or DR5 knockdown cells to GODZ and TRAIL. HeLa cells were stably transfected with control pshRNA (Ctrl), pDR5 shRNA (shDR5) (e), or pDR4 shRNA (shDR4) (f), and their expression levels were examined by western blotting using anti-DR4, anti-DR5, and anti-β-actin antibodies (inserts). HeLa stable cells were then transfected with pEGFP (Clontech), and either pcDNA3-HA (Ctrl) or pGODZ-HA (GODZ) for 24 h and then left untreated (MOCK) or exposed to TRAIL (100 ng/ml) for 3 h. Cell death ratios were measured as described in (b). Bars indicate mean±S.D. (n=3). P-values were estimated using t-test versus control
Contribution of GODZ DHHC motif to PM targeting of DR4 and TRAIL sensitivity
DHHC-containing protein family is implicated for their role in the membrane targeting or trafficking of diverse substrates. Given that GODZ contains DHHC motif and DR4 is targeted to the PM, we first addressed whether the DHHC motif of GODZ regulated TRAIL-induced apoptosis. We introduced mutations into the DHHC motif of GODZ to generate GODZ-C157S and GODZ-H155A mutants, which replaced Cys157 with Ser and His155 with Ala, respectively. Unlike wild-type GODZ, ectopic expression of GODZ-C157S or GODZ-H155A mutant in HeLa cells was neither effective to increase TRAIL-induced apoptosis (Figure 5a) nor affected biochemical profiles, such as caspase activation, of TRAIL-induced apoptosis (Figure 5b), suggesting that the DHHC motif of GODZ is functionally required for TRAIL-induced apoptosis. From immunoprecipitation analysis, however, we found that GODZ-C157S mutant interacted with DR4 as much as wild-type GODZ (Supplementary Figure S3a).
Contribution of GODZ DHHC motif to plasma membrane (PM) targeting of DR4 and TRAIL sensitivity. (a) Mutation in GODZ DHHC motif loses the ability to stimulate TRAIL-mediated cell death. HeLa cells were transiently co-transfected with pEGFP (Clontech) and either pcDNA3-HA (Ctrl), pGODZ WT-HA, pGODZ(C157S)-HA, or pGODZ(H155A)-HA for 24 h, and then left untreated (MOCK) or treated with TRAIL (100 ng/ml) for the indicated times. After staining with EtHD, cell death was measured by counting the number of both GFP and EtHD-positive cells among total GFP-positive cells. Bars indicate mean±S.D. (n=3). P-values were estimated using t-test versus control (*P<0.05; **P<0.01; ***P<0.001). (b) Lack of caspase-8 activation by GODZ mutants. HeLa cells were transiently transfected with pcDNA3-HA, pGODZ-HA, pGODZ(C157S)-HA, or pGODZ(H155A)-HA for 24 h, and left untreated or treated with TRAIL (100 ng/ml) for 3 h. Cell extracts were prepared and analyzed by western blotting using the indicated antibodies. (c) Mutation in cysteine-rich motif of DR4 (DR4C261-3S) weakens the pro-apoptotic activity. HeLa cells were transfected with pcDNA3 (PCD; an empty vector) or pGODZ, and pEGFP (Clontech) (Ctrl), pDR4-GFP, or pDR4 (C261-3S)-GFP for 18 h, and cell death ratios were measured as described in (a). Bars indicate mean±S.D. (n=3). P values were estimated using t-test versus control (*P<0.05; **P<0.01). (d) Failure in the targeting of DR4 C261-3S mutant to PM. COS7 cells were transiently transfected with pDR4-GFP or pDR4 (C261-3S)-GFP for 12 h. Thereafter, cells were fixed and stained with anti-GM130 antibody (red). Green and red fluorescent images were acquired under a confocal microscope (Zeiss, Thronwood, NY, USA) and then overlaid with Hoechst 33258 staining. (e) Fractionation assay showing the impaired targeting of DR4 C261-3S mutant to PM. HEK 293T cells were transiently transfected with pDR4-HA or pDR4 (C261-3S)-HA for 24 h, and then cell extracts were subjected to fractionation assay to separate the PM from the cytosol (C), as described in materials and methods. The fractions were analyzed by western blotting with anti-HA and anti-β-actin antibodies. (f) Effects of various DHHC motif-containing proteins on TRAIL sensitivity. HeLa cells were transiently transfected with pEGFP (Clontech) and various DHHC motif-containing constructs for 24 h, and then left untreated (MOCK) or exposed to TRAIL (100 ng/ml) for 3 h. Cell death ratios were measured as described in (a). Bars indicate mean±S.D. (n=3). P-values were estimated using t-test versus control (*P<0.05; **P<0.01; ***P<0.001). The color reproduction of this figure is available at the Cell Death and Differentiation journal online
A recent report showed that DR4 is modified by palmitoylation at its cysteine-rich motif, and DR4 C261-3S mutant that has replaced Cys261-263 with Ser and is palmitoylation-defective, is less efficient in transmitting a cell death.30 By using DR4 C261-3S mutant, we addressed functional interaction for the possible palmitoylation between GODZ and DR4. Unlike wild-type DR4, overexpression of DR4 C261-3S mutant itself was not much active to induce cell death (Figure 5c) and was not effective to increase TRAIL-induced cell death in HeLa cells (Supplementary Figures S4a and b). Moreover, coexpression with GODZ did not sensitize cell death mediated by DR4 C261-3S mutant (Figure 5c). There was no difference in the expression levels between DR4 and DR4 C261-3S mutant (Figure 5c, insert). Interestingly, from confocal image analysis using GFP-fused constructs, we observed that DR4 C261-3S-GFP apparently accumulated in the golgi apparatus and irregularly diffused in the cytosol, whereas DR4-GFP was located mainly in the PM (Figure 5d). Using alternative fractionation analysis, we detected DR4 in both the PM and cytosol fractions of HEK 293T cells, whereas DR4 C261-3S mutant was exclusively in the cytosol fraction (Figure 5e). All together, these data demonstrate that GODZ may regulate cell surface expression of DR4 via cysteine-rich motif in the C-terminus, and is thus crucial for DR4-mediated apoptotic signaling upon TRAIL engagement. Furthermore, among DHHC motif-containing proteins-2, -3, -4, -8, and -9, GODZ was most efficient to enhance TRAIL sensitivity (Figure 5f), adding more specificity to the role of GODZ in TRAIL signaling. Expressions of DHHC cDNAs were observed by GFP-positive signals under a fluorescence microscope (Supplementary Figure S3b) and were detected by western blot analysis (Supplementary Figure S3c).
Because DHHC motif of GODZ has a potency to target DR4 to the PM, we further assessed a role of GODZ in cell surface expression of endogenous TRAIL receptors. After reduction of GODZ expression using shRNAs (GODZ shRNA #1, and #2), we performed biotinylation assay using Sulfo-NHS-SS-Biotin (Pierce biotechnology, Rockford, IL, USA). Western blot analysis showed that cell surface localization of DR4 was markedly reduced in HeLa/GODZ knockdown cells (Figure 6a). On the other hand, cell surface localization of DR5 was not affected in the same cells. In addition, we investigated the distinct subcellular distribution of DR4 using discontinuous gradient centrifugation (Figures 6b and c). In control cells, we detected DR4 and DR5 in the high-density fraction containing the golgi apparatus, which includes GM130, a golgi marker, and the ER that contains GRP78, an ER marker, and also in the low-density fraction containing PM (Figure 6b). In HeLa/GODZ knockdown cells, the amount of DR4 detected in the low-density fraction was apparently decreased but concomitantly increased in the high-density fraction, whereas DR5 was not changed. The quantification of these changes in the PM pools containing the fractions 7, 8, and 9 with western blot analysis clearly showed that cell surface localization of DR4 was impaired in HeLa/GODZ knockdown cells (Figure 6c). Conversely, overexpressed GODZ enhanced cell surface localization of DR4, whereas GODZ-C157S mutant failed to do so, as assessed with biotinylation assay (Figure 6d). In addition, we found that GODZ-A mutant, which interacted with DR4 but could not sensitize TRAIL-mediated apoptosis, failed to deliver DR4 to the cell surface (Figure 6e). Thus, GODZ appears to primarily regulate the PM localization of DR4, but not DR5.
Depletion of GODZ expression reduces PM-localized DR4. (a) Biotinylation assay showing GODZ-dependent localization of DR4 to the PM. HeLa/Ctrl shRNA and HeLa/GODZ shRNA stable cells were exposed to biotin, and the PM fractions were prepared by fractionation and subjected to biotinylation assays with Sulfo-NHS-SS-Biotin (Pierce biotechnology). Western blotting was then performed with anti-GODZ, anti-DR4, anti-DR5, and anti-β-actin antibodies. Densitometric analysis of western blots was performed using a multi-gauge imaging system and their relative ratios to that of control shRNA are represented. (b and c) Fractionation assay using gradient centrifugation shows impaired membrane targeting of DR4 by GODZ deficiency. HeLa/Ctrl shRNA and HeLa/GODZ shRNA cell extracts were subjected to discontinuous gradient centrifugation using OptiPrep (Sigma) (iodixanol) as a gradient medium. After centrifugation, nine fractions of equal volume (500 μl each) were concentrated and subjected to western blotting using anti-DR4, anti-DR5, anti-GRP78, and anti-GM130 antibodies (b). The fractions seven, eight, and nine were pooled and subjected to western blotting using anti-DR4, anti-DR5, and anti-PARP-1 antibodies. Densitometry analysis of western blots was performed and relative ratios of DR4 and DR5 signals to control are represented (c). (d) Enhanced targeting of DR4 to cell surface by GODZ but not by GODZ C157S mutant. HEK 293T cells were transfected with pDR4-Flag and either pGODZ-HA or pGODZ(C157S)-HA for 24 h, and then subjected to biotinylation assay with (Pierce biotechnology) as described in (a). Western blotting was performed with anti-Flag and anti-HA antibodies. (e) Effects of GODZ mutants on targeting of DR4 to PM. HEK 293T cells were co-transfected with pDR4-Flag and either pGODZ-HA or GODZ deletion mutant for 24 h. The PM and cytosol fractions were prepared as described in materials and methods, and subjected to western blotting using anti-Flag or anti-HA antibody. The asterisk in (e) indicates nonspecific signal of the cytosol fraction. HF, high-density fraction; LF, low-density fraction; P, pallet; T, total
Reconstitution of GODZ expression sensitizes Hep-3B tumor cells to TRAIL
Previous studies showed that the GODZ protein is broadly expressed in many tissues, including liver.25, 31 When we examined the expression of GODZ in normal and cancer cell lines, we found that GODZ expression was detected in almost all of the cells, including HEK 293T cells and HeLa cells (Figure 7a). Further, we found that the expression of DR4 was low in TRAIL-resistant HEK 293T cells and high in TRAIL-sensitive HeLa cells. Interestingly, Hep-3B human hepatocellular carcinoma cells showed high expression of DR4 but very low levels of GODZ and DR5. From apoptosis assay using TRAIL, we found that Hep-3B cells were most resistant to TRAIL among those cells we tested. We then hypothesized that Hep-3B cells might be insensitive to TRAIL in spite of high expression of DR4 because of the impaired subcellular localization of DR4 to cell surface, which may result from low level of GODZ expression.
Reconstitution of GODZ expression sensitizes Hep-3B tumor cells to TRAIL with more DR4 in the PM. (a) Expression analysis of TRAIL receptors and GODZ in tumor cells and TRAIL-sensitivity. HEK 293T, HeLa, A549, Hep-3B, MCF7, and COS7 cells were collected and examined for their sensitivities to TRAIL. Cell extracts were prepared and subjected to western blotting using anti-DR4, anti-DR5, anti-GODZ, and anti-β-actin antibodies (left). The summary of these results is represented by the plus (+) for high expression and their sensitivity to TRAIL (right). (b and c) Reconstitution of GODZ sensitizes Hep-3B cells to TRAIL. Hep-3B cells were transiently transfected with pEGFP (Clontech) and either pcDNA3-HA (Ctrl) or pGODZ-HA for 24 h, and then left untreated (MOCK) or treated with TRAIL (100 ng/ml) for 12 h in the presence or absence of caspase inhibitor, IDN6556. Bars indicate mean±S.D. (n=3). P values were estimated using t-test versus control (b). Cell extracts were prepared and examined by western blotting using anti-caspase-8, anti-HA, and β-actin antibodies (c). (d) Reconstitution of GODZ enhances PM localization of DR4. Hep-3B cells were transiently transfected with pcDNA3-HA or pGODZ-HA for 24 h. Cell extracts were subjected to fractionation analysis to prepare PM fraction. The fractions and WCL were analyzed by western blotting using the indicated antibodies
Thus, we reconstituted Hep-3B cells with GODZ and examined their sensitivity to TRAIL. Ectopic expression of GODZ increased the sensitivity of Hep-3B cells to TRAIL about twofold (Figure 7b). We also observed that proteolytic activation of caspase-8 was significantly increased in Hep-3B cells after treatment with TRAIL following reconstitution with GODZ, which was blocked by pan-caspase inhibitor (Figure 7c). More importantly, western blot analysis following fractionation assay showed that compared with control cells, cell surface localization of DR4 was significantly increased in Hep-3B cells, which were reconstituted with GODZ-HA (Figure 7d). On the other hand, DR5 was not affected. Thus, these results suggest that expressional regulation of GODZ may contribute to cancer surveillance in TRAIL-resistant cells by acting as a determinant in the targeting of TRAIL receptor, DR4, to cell surface.
Discussion
In human, DR4 seems to share common death machineries with DR5, forming DISC complex to activate proximal caspases for apoptosis by TRAIL. However, the potential difference in DR4- versus DR5-mediated apoptosis is not clear. Unlike DR5, increasing evidence showed that DR4 is distinctly regulated at multilevels in spite of high sequence similarity (∼65%) with death domains of DR5.11 The advent of SOS protein recruitment–yeast two-hybrid assay enabled us to excavate a membrane-bound regulator of DR4, thus unveiling the necessity of GODZ in targeting of DR4 to the PM for DR4-mediated signaling. Similarly, cell surface expression of TRAIL receptors can be differentially regulated by signal recognition particle (SRP) complex and ARAP1 at the posttranslational level.32, 33 In addition, we also previously found that ribosomal protein S6 selectively regulates DR4 expression, and thus TRAIL sensitivity of tumor cells.20 Moreover, the expression of DR4 is exclusively regulated by genotoxic stress such as ionizing radiation34 and combinational treatments with DNA-damaging agent.15 These observations implicate the distinct role of DR4 in TRAIL signaling.
Previous studies suggest that DR4 may have an important role in some tumor cells, including chronic lymphocytic leukemia (CLL), ovarian, and breast cancers in which the cell death by TRAIL is predominately signaled by DR4.35, 36 TRAIL can alter their inclination to activate different death receptors through selective regulator such as GODZ and SRP. We found that DR4 is expressed but not properly located into the cell surface of liver tumor cells. In GODZ-depleted cancer cells, DR4 appeared to accumulate in the ER/golgi fractions with losing in the cell surface, whereas DR5 localization and function were unaffected. Previous report showed that the SRP complex also has an essential role in the localization of DR4 from ER to the cell surface expression.33 We speculate that SRP and GODZ may have roles together in the concerted pathway; SRP probably has a role in targeting of DR4 from ER to golgi and GODZ, then from golgi apparatus to the PM for proper localization. Thus, dysregulation of these modulators may provide a hint to understand how cancer cells acquire the resistance to TRAIL in spite of normal expression of TRAIL receptor.
Recently, it was reported that the palmitoylation of DR4 is required for the localization into lipid rafts, and therefore for an efficient transmission of apoptotic signal.30 In our study, a DR4 mutant defective in the potential palmitoylation was barely observed in the cell surface and was not effective to induce cell death. Also, GODZ mutant in the DHHC motif failed to function to target DR4 to the PM. Based on these findings, we believe that GODZ, holding PAT activity, binds to DR4 to facilitate cell surface localization of DR4, probably through the palmitoylation of DR4. It will be interesting to test whether mouse GODZ can regulate targeting of murine TRAIL receptor to the PM. Recently, GODZ was reported to increase protein affinity of RGS4 to cell membranes in breast cancer cells.37 In addition, the DHHC region and C-terminal transmembrane domains of GODZ were needed for its binding to DR4. But, these regions failed to show the ability of targeting DR4 to the cell surface, and thus transducing the apoptosis signal. Thus, the N-terminus of GODZ, as well as the DHHC and C-terminus, may be required for the enzyme activity and targeting DR4 to the membrane. It will also be interesting to map the region of GODZ responsible for interacting with and targeting GODZ substrates, including NMDA receptor, to the cell surface of neuronal cells.28
Despite that TRAIL can induce apoptosis selectively with potent antitumor activity in a wide variety of cancer cells, it has been reported that several types of cancer cells are resistant to TRAIL-induced apoptosis. Diverse mechanisms have been suggested by which individual cancer cells overcome their sensitivities to TRAIL. Defect in cell surface expression of death receptors appears to be sufficient to determine the fate of cancer exposed to chemotherapeutic ligands. Vice versa, all with similar cell surface levels of DR4 and DR5, DR5 seems more important in lung, colon carcinoma, and melanoma.38, 39 However, the reason by which some cancer cells respond to TRAIL preferentially via either DR4 or DR5 has not been established. Thus, it will be interesting to examine the expression level of GODZ and targeting of DR4 to the PM in these cancers, as we found that GODZ was downregulated in Hep-3B hepatocarcinoma cells showing TRAIL-resistance. Hep-3B cells expressed relatively low level of DR5 and were unaffected by TRAIL. Our observation that reconstitution with GODZ increased cell surface expression of DR4 and also tumor cells’ sensitivity to TRAIL, providing insight in TRAIL-resistance in tumors and TRAIL therapeutics.
Taken together, our study suggests that GODZ facilitates targeting of DR4 to the PM through DHHC motif, and thereby enhances TRAIL-mediated apoptosis in tumor cells, postulating GODZ as a TRAIL and DR4-selective regulator.
Materials and Methods
DNA constructs
The cDNA of cytoplasmic domain (273–468) in DR4 was amplified by PCR using synthetic oligonucleotides (forward 5′-CGG GAT CCC GCC CAA GTG CAT GGA CAG G-3′ and reverse 5′-CAT TTA GGT GAC ACT A-3′). The amplified product was subcloned into BamHI site of pSOS (pSOS-DR4) system (Stratagene, La Jolla, CA, USA). The full cDNA of human GODZ was amplified by PCR using synthetic oligonucleotides (forward 5′-CCC AAG CTT ATG ATG CTT ATC CCC ACC CAC CAT TCC GAA ACA TTG AG-3′ and reverse 5′-CGG GAT CCC GGA CCA CAT ACT GGT ACG G-3′, and forward 5′-CGC TCT AGA ATG CTT ATC CCC ACC-3′ and reverse 5′-CGC TCT AGA TCA GAC CAC ATA CTG GTA C-3′) and the PCR products were subcloned into pcDNA3-HA (Invitrogen, Carlsbad, CA, USA). Deletion constructs of GODZ were amplified by PCR and subcloned into pcDNA3-HA. Human DHHC (2, 3, 4, 8, and 9) cDNAs were subcloned into GFP-DEST to generate GFP fusion proteins using GATEway recombination system (Invitrogen). PCR products of human DR4 and DR5 cDNA were subcloned into pcDNA3-HA, p3xFLAG-CMV (Sigma, St. Louis, MO, USA), and pEGFP-N1 (Clontech, Palo Alto, CA, USA). The GODZ shRNAs were constructed using forward and reverse synthetic 19-nucleotides (#1, 331-AGG AAA TGC CAC TAA AGA A-339, #2, 504-CCA GAA GTA CTT CGT CCT G-522), which were synthesized, annealed, and cloned into BglII and HindIII sites of pSuper.neo (OligoEngine, Seattle, WA, USA). The pDR4 shRNAs were generated as previously described.20 The DR5 shRNA (#1, 108-GAC CCT TGT GCT CGT TGT C-126, #2, 1097-ATG AGA TAA AGG TGG CTA A-1115) were constructed as described above.
Cell culture and DNA transfection
HeLa and HEK 293T cells were cultured in DMEM (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (HyClone), penicillin, and streptomycin (Invitrogen). Hep-3B cells were maintained in RPMI 1640 (HyClone) supplemented with 10% FBS. Cells were cultured at 37 °C under an atmosphere of 5% CO2. Transfection was carried out using LipofectAMINE (Invitrogen), Polyfect reagent (Qiagen, Valencia, CA, USA), or PEI (Sigma) according to the manufacturers’ instructions. Typically, 2 × 105 cells per well in 6-well dishes were transfected with appropriate plasmids.
Site-directed mutagenesis
DR4 (261-3S), GODZ (C157S), and GODZ (H155A) mutants were generated by a Quickchange Site-Directed Mutagenesis Kit (Stratagene) using synthetic oligonucleotides containing mutations in the corresponding positions. All mutants were verified by DNA sequencing analysis.
Cell death assay
Cells were exposed to various stimuli and double-stained with Hoechst 33258 (1 μg/ml) and ethidium homodimer (EtHD) (Molecular Probes, Eugene, OR, USA), after which apoptotic cell death was assessed by counting the number of GFP-positive cells showing condensed and fragmented nuclei under a fluorescence microscope (Olympus, Tokyo, Japan).
Generation of stable cell lines
HeLa cells were transfected with pcDNA3-HA, pGODZ-HA, or pGODZ shRNA for 24 h and then grown in the selection medium containing 1 mg/ml of G418 (Invitrogen) for 2 weeks. Then, single clones of stable cells were isolated by the standard way and analyzed by western blotting.
BiFC plasmids and assay
Human GODZ cDNA was fused to the N-terminal Venus fragment of pBiFC-VN (pGODZ-VN) and Human DR4 and DR5 were fused to the C-terminal Venus fragments of pBiFC-VC (pDR4-VC and pDR5-VC, respectively). BiFC assay was performed as described in the text.21, 29 HeLa cells were transfected with VN or VC-fusion construct alone or in combination, and incubated at 37 °C for 19 h. After staining cell nuclei with Hoechst 33258, fluorophore formation in the living cells was imaged using a fluorescence microscope (Olympus).
Antibodies and western blotting
Anti-PARP-1 (sc-8007), anti-GODZ (sc-79960), anti-β-actin (sc-47778) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-DR4 (#13890), anti-DR5 (Abcam, Cambridge, MA, USA; #47179), anti-Flag (M2) (Sigma; F3165), anti-GM130 (BD Biosciences, CA, USA; #610823), anti-caspase-10 (MBL, M059-3), anti-caspase-9 (Cell Signaling, Danvers, MA, USA; #9502), and anti-caspase-8 were used in the analysis. In general, cells were lyzed in ice-cold RIPA buffer (50 mM Tris-Cl pH 8.0, 15 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin A) and sonicated briefly. Cell lysates were clarified by centrifugation, resolved by SDS-PAGE, and proteins on gels were transferred to nitrocellulose (Millipore, Billerica, MA, USA) using a semidry blotter (Bio-Rad, Hercules, CA, USA). The membrane was blocked with 3% BSA in TBST (10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% Tween-20) and incubated overnight at 4 °C with primary antibodies. Immunoblots were visualized by enhanced chemiluminescence method. To detect the endogenous GODZ, cells were lyzed in ice-cold CHAPS buffer (30 mM Tris-Cl pH 7.5, 150 mM NaCl, 1% CHAPS, 1 mM PMSF, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin A).
Immunoprecipitation assay
For endogenous immunoprecipitation assay, 2 × 107 HeLa cells were lyzed in 700 μl CHAPS buffer containing 30 mM Tris-Cl pH 7.5, 150 mM NaCl, 1% CHAPS, 1 mM PMSF, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin A. The cell lysates were incubated for 12 h at 4 °C with polyclonal anti-GODZ antibody or pre-immune serum after preclearance with 20 μl protein A/G–Bead (Santa Cruz Biotechnology). For co-immunoprecipitation assay, 2 × 106 HEK 293T cells were transfected with Flag-tagged DRs and HA-tagged GODZ, and lyzed in ice-cold RIPA buffer (50 mM Tris-Cl pH 8.0, 15 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, and 1 μg/ml each of aprotinin, leupeptin, and pepstatin A). The cell lysates were incubated for 4 h at 4 °C with monoclonal HA or Flag antibody. After adding another 30 μl protein A/G–Bead, the mixtures were incubated for an additional 3 h at 4 °C. The immunocomplexes were collected by centrifugation at 5000 r.p.m. for 3 min, washed four times with lysis buffer, and detected by western blotting, as described above.
Immunostaining
COS7 cells grown on coverslips in 12-well plates were fixed in 4% paraformaldehyde for 5 min, permeabilized with 0.1% Triton X-100 for 5 min, and blocked with 1% BSA for 2 h. After stained with antibody and Hoechst 33258, cells were visualized under a LSM confocal fluorescence microscope (Zeiss, Thronwood, NY, USA).
RT-PCR
Total RNA was purified from HeLa cells using MRC Trizol reagent (Molecular Research Center, Inc., Cincinnati, OH, USA) and subjected to amplification with SuperScript One-Step RT-PCR system (Invitrogen). The following specific primers sets were used for PCR; GODZ, forward 5′-ATG ATG CTT ATC CCC ACC-3′ and reverse 5′-TCA GAC CAC ATA CTG GTA-3′; DR4, forward 5′-ATG GCG CCA CCA CCA GCT-3′ and reverse 5′-TCA CTC CAA GGA CAC GGC-3′; actin, forward 5′-ATG GGG AAG GTG AAG GTC GGA-3′ and reverse 5′- CCA TGA CGA ACA TGG GGG CAT-3′.
Biotinylation assay
Biotinylation assay was carried out using Sulfo-NHS-SS-Biotin (Pierce biotechnology) according to the manufacturer's instructions. Briefly, cell lysates were washed three times with ice-cold PBS and incubated with Sulfo-NHS-SS-Biotin (Pierce biotechnology, 0.15 mg/ml) in PBS for 1 h at 4 °C. After washing cells three times with ice-cold PBS (pH 8.0) containing glycine to remove non-reacted biotinylation reagent, cells were lyzed in ice-cold RIPA buffer. After centrifugation (16 000 × g for 15 min at 4 °C), supernatants containing equal amount of protein were incubated with streptavidin beads to immunoprecipitate the remaining biotinylated proteins. The immunocomplexes were collected by centrifugation at 5000 r.p.m. for 3 min, washed four times with lysis buffer, and detected by western blotting, as described above.
Subcellular fractionation
Cells grown in 100 mm culture plates were harvested with buffer containing 0.25 M sucrose, 1 mM EDTA, 10 mM HEPES-NaOH pH 7.4 and then mechanically disrupted by a 26-gauge needle with passing it 20 times. The cell suspension was centrifuged at 1000 × g for 10 minutes at 4 °C to remove the nuclei, and the supernatant was carefully aspirated and again spun at 65 000 × g for 60 min at 4 °C with Beckman SW41 type rotor. The precipitate was diluted with a OptiPrep (Sigma) to final concentration from 30 to 2.5% using diluent (0.25 M sucrose, 6 mM EDTA, 60 mM HEPES-NaOH pH 7.4) and carefully layered sequentially on top of the supernatant, and the tube was spun at 100 000 × g for 16 h at 4 °C using the swinging-bucket rotor without the brake. Finally, gradient in 0.5 ml fractions were collected.
Preparation of cytosol/PM fractions
Fractionation assay was performed as described.40 Briefly, to prepare the PM and cytosol fractions, cultured cells were harvested with buffer A (50 mM Tris-Cl, pH 8.0, and 0.5 mM DTT containing 0.1% (v/v) NP-40, protease and phosphatase inhibitors). After homogenation, the lysates were sheared by a 26-gauge needle with passing it seven times. Each lysates were centrifuged at 1000 × g for 10 min at 4 °C, and the precipitate obtained was suspended in NP-40-free buffer A, incubated on ice for 10 min with occasional mixing, and centrifuged at 1000 × g for 10 min at 4 °C. The precipitates were suspended again in buffer A containing 1% (v/v) NP-40, incubated on ice with occasional mixing, and centrifuged at 16 000 × g for 1 h at 4 °C. The supernatants were collected and stored as the PM fraction at −70 °C. The supernatants from the first and second centrifugation were gathered and centrifuged at 16 000 × g for 1 h at 4 °C. The supernatant was used as a cytosol fraction.
Cytotrap method
The two-hybrid screening was conducted using the CytoTrap system (Stratagene) according to the manufacturer's instructions. Yeast cdc25H contains a point mutation at amino-acid residue 132 of the cdc25 gene, the yeast homolog of human SOS gene. It prevents host growth at 37 °C, but host growth is normal at the permissive temperature (25 °C). Yeast cdc25H was transformed with pSOS-DR4 (amino acids 246–445) (Stratagene) and a Human thymus cDNA library cloned into the pMyr vector (Stratagene). A total of 7 × 105 clones were analyzed. Transformed yeast was selected on SD/glucose agar plate lacking uracil and leucine at 25 °C for 2 days. Transformed yeast was selected on SD/galactose/raffinose agar plate lacking uracil and leucine at 37 °C for 6–10 days. The colonies that grow at 37 °C were blotted onto SD/galactose/raffinose lacking uracil and leucine and SD/glucose lacking uracil and leucine, which were selected at 37 °C for 5 days. As a negative control, yeast expressing only bait was spreaded on SD/glucose/-Ura/-Leu plate.
TRAIL preparation
Recombinant TRAIL preparation was described previously.4
Statistical analysis
Data are expressed as mean±S.D. Statistical comparisons between groups were performed using one-way analysis of variance (ANOVA) followed by Bonferroni's test or by the Student's t-test as appropriate. Probabilities of P<0.05 were considered statistically significant.
Abbreviations
- BiFC:
-
bimolecular fluorescence complementation
- CRD:
-
cysteine-rich domain
- DR:
-
death receptor and TRAIL-R
- DHHC:
-
Asp-His-His-Cys
- DISC:
-
death-inducing signaling complex
- EtHD:
-
ethidium homodimer
- GODZ:
-
Golgi-specific DHHC Zinc Finger Protein
- TRAIL:
-
tumor necrosis factor-related apoptosis-inducing ligand
References
Ashkenazi A, Dixit VM . Death receptors: signaling and modulation. Science 1998; 281: 1305–1308.
Kim K, Fisher MJ, Xu SQ, el-Deiry WS . Molecular determinants of response to TRAIL in killing of normal and cancer cells. Clin Cancer Res 2000; 6: 335–346.
Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY et al. Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev 2003; 17: 883–895.
Kim IK, Chung CW, Woo HN, Hong GS, Nagata S, Jung YK . Reconstitution of caspase-8 sensitizes JB6 cells to TRAIL. Biochem Biophys Res Commun 2000; 277: 311–316.
Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A . Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 2000; 12: 611–620.
Lin Y, Devin A, Cook A, Keane MM, Kelliher M, Lipkowitz S et al. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IkappaB kinase and c-Jun N-terminal kinase. Mol Cell Biol 2000; 20: 6638–6645.
Schneider P, Thome M, Burns K, Bodmer JL, Hofmann K, Kataoka T et al. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-kappaB. Immunity 1997; 7: 831–836.
Wu GS, Burns TF, Zhan Y, Alnemri ES, El-Deiry WS . Molecular cloning and functional analysis of the mouse homologue of the KILLER/DR5 tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor. Cancer Res 1999; 59: 2770–2775.
Finnberg N, Klein-Szanto AJ, El-Deiry WS . TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis. J Clin Invest 2008; 118: 111–123.
Fulda S, Debatin KM . Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene 2006; 25: 4798–4811.
LeBlanc HN, Ashkenazi A . Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ 2003; 10: 66–75.
Sedger LM, Shows DM, Blanton RA, Peschon JJ, Goodwin RG, Cosman D et al. IFN-gamma mediates a novel antiviral activity through dynamic modulation of TRAIL and TRAIL receptor expression. J Immunol 1999; 163: 920–926.
Zhang X, Inukai T, Akahane K, Hirose K, Kuroda I, Honna H et al. Endoplasmic reticulum stress inducers, but not imatinib, sensitize Philadelphia chromosome-positive leukemia cells to TRAIL-mediated apoptosis. Leukemia Res 2011; 35: 940–949.
Liu X, Yue P, Khuri FR, Sun SY . p53 upregulates death receptor 4 expression through an intronic p53 binding site. Cancer Res 2004; 64: 5078–5083.
Mendoza FJ, Ishdorj G, Hu X, Gibson SB . Death receptor-4 (DR4) expression is regulated by transcription factor NF-kappaB in response to etoposide treatment. Apoptosis 2008; 13: 756–770.
Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science 1997; 277: 818–821.
Kim Y, Suh N, Sporn M, Reed JC . An inducible pathway for degradation of FLIP protein sensitizes tumor cells to TRAIL-induced apoptosis. J Biol Chem 2002; 277: 22320–22329.
Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG . A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science 2004; 305: 1471–1474.
Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med 2007; 13: 1070–1077.
Jeon YJ, Kim IK, Hong SH, Nan H, Kim HJ, Lee HJ et al. Ribosomal protein S6 is a selective mediator of TRAIL-apoptotic signaling. Oncogene 2008; 27: 4344–4352.
Choi H, Chang JW, Jung YK . Peroxiredoxin 6 interferes with TRAIL-induced death-inducing signaling complex formation by binding to death effector domain caspase. Cell Death Differ 2011; 18: 405–414.
Sharma C, Yang XH, Hemler ME . DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151. Mol Biol Cell 2008; 19: 3415–3425.
Zhang J, Planey SL, Ceballos C, Stevens Jr SM, Keay SK, Zacharias DA . Identification of CKAP4/p63 as a major substrate of the palmitoyl acyltransferase DHHC2, a putative tumor suppressor, using a novel proteomics method. Mol Cell Proteomics 2008; 7: 1378–1388.
Ducker CE, Stettler EM, French KJ, Upson JJ, Smith CD . Huntingtin interacting protein 14 is an oncogenic human protein: palmitoyl acyltransferase. Oncogene 2004; 23: 9230–9237.
Uemura T, Mori H, Mishina M . Isolation and characterization of Golgi apparatus-specific GODZ with the DHHC zinc finger domain. Biochem Biophys Res Commun 2002; 296: 492–496.
Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoe-Pognetto M et al. The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J Neurosci 2004; 24: 5881–5891.
Fang C, Deng L, Keller CA, Fukata M, Fukata Y, Chen G et al. GODZ-mediated palmitoylation of GABA(A) receptors is required for normal assembly and function of GABAergic inhibitory synapses. J Neurosci 2006; 26: 12758–12768.
Hayashi T, Rumbaugh G, Huganir RL . Differential regulation of AMPA receptor subunit trafficking by palmitoylation of two distinct sites. Neuron 2005; 47: 709–723.
Hu CD, Chinenov Y, Kerppola TK . Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 2002; 9: 789–798.
Rossin A, Derouet M, Abdel-Sater F, Hueber AO . Palmitoylation of the TRAIL receptor DR4 confers an efficient TRAIL-induced cell death signalling. Biochem J 2009; 419: 185–192.
Hines RM, Kang R, Goytain A, Quamme GA . Golgi-specific DHHC zinc finger protein GODZ mediates membrane Ca2+ transport. J Biol Chem 2010; 285: 4621–4628.
Simova S, Klima M, Cermak L, Sourkova V, Andera L . Arf and Rho GAP adapter protein ARAP1 participates in the mobilization of TRAIL-R1/DR4 to the plasma membrane. Apoptosis 2008; 13: 423–436.
Ren YG, Wagner KW, Knee DA, Aza-Blanc P, Nasoff M, Deveraux QL . Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol Biol Cell 2004; 15: 5064–5074.
Di Pietro R, Secchiero P, Rana R, Gibellini D, Visani G, Bemis K et al. Ionizing radiation sensitizes erythroleukemic cells but not normal erythroblasts to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)–mediated cytotoxicity by selective up-regulation of TRAIL-R1. Blood 2001; 97: 2596–2603.
MacFarlane M, Inoue S, Kohlhaas SL, Majid A, Harper N, Kennedy DB et al. Chronic lymphocytic leukemic cells exhibit apoptotic signaling via TRAIL-R1. Cell Death Differ 2005; 12: 773–782.
Horak P, Pils D, Haller G, Pribill I, Roessler M, Tomek S et al. Contribution of epigenetic silencing of tumor necrosis factor-related apoptosis inducing ligand receptor 1 (DR4) to TRAIL resistance and ovarian cancer. Mol Cancer Res 2005; 3: 335–343.
Wang J, Xie Y, Wolff DW, Abel PW, Tu Y . DHHC protein-dependent palmitoylation protects regulator of G-protein signaling 4 from proteasome degradation. FEBS Lett 2010; 584: 4570–4574.
Lee SH, Shin MS, Kim HS, Lee HK, Park WS, Kim SY et al. Alterations of the DR5/TRAIL receptor 2 gene in non-small cell lung cancers. Cancer Res 1999; 59: 5683–5686.
Laguinge LM, Samara RN, Wang W, El-Deiry WS, Corner G, Augenlicht L et al. DR5 receptor mediates anoikis in human colorectal carcinoma cell lines. Cancer Res 2008; 68: 909–917.
Nishiumi S, Ashida H . Rapid preparation of a plasma membrane fraction from adipocytes and muscle cells: application to detection of translocated glucose transporter 4 on the plasma membrane. Biosci Biotechnol Biochem 2007; 71: 2343–2346.
Acknowledgements
We thank Dr TK Kerppola for BiFC plasmids. Mr Y Oh was partially supported by BK21 program. This work was supported by the Grants from the Global Research Laboratory program (K21004000002-11A0500-00210) and the BAERI project (2011-0006314) funded by the Ministry of Education, Science and Technology (MEST) and the Cancer project (0820120) funded by the Ministry of Human Healthy and Welfare in Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by H Ichijo
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Rights and permissions
About this article
Cite this article
Oh, Y., Jeon, YJ., Hong, GS. et al. Regulation in the targeting of TRAIL receptor 1 to cell surface via GODZ for TRAIL sensitivity in tumor cells. Cell Death Differ 19, 1196–1207 (2012). https://doi.org/10.1038/cdd.2011.209
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2011.209
Keywords
This article is cited by
-
Cardioprotective role of APIP in myocardial infarction through ADORA2B
Cell Death & Disease (2019)
-
Identification of PTPN1 as a novel negative regulator of the JNK MAPK pathway using a synthetic screening for pathway-specific phosphatases
Scientific Reports (2017)
-
Fas palmitoylation by the palmitoyl acyltransferase DHHC7 regulates Fas stability
Cell Death & Differentiation (2015)
-
TRAIL and vitamins: opting for keys to castle of cancer proteome instead of open sesame
Cancer Cell International (2012)