Abstract
Background:
Recently several microRNAs (miRNAs) have been found to be regulated by genistein in cancer cells. In this study, we focused on the gene regulatory effect of genistein on microRNA and its target genes in prostate cancer (PC).
Methods:
Initially, we investigated the effect of genistein on prostate cancer cells and identified that the expression of miRNA-1260b was decreased by genistein. We performed functional analyses and investigated the relationship between miRNA-1260b expression and prostate cancer patient outcomes. Two target genes (sFRP1 and Smad4) of miR-1260b were identified based on computer algorithm and 3′UTR luciferase assay was carried out to determine direct miRNA regulation of the genes.
Results:
Genistein promoted apoptosis while inhibiting prostate cancer cell proliferation, invasion and TCF reporter activity in PC cells. MiR-1260b was highly expressed in prostate cancer tissues and significantly downregulated by genistein in PC cells. After knocking down miR-1260b, cell proliferation, invasion, migration and TCF reporter activity were decreased in PC cells. Western analysis and 3′UTR luciferase assay showed that the two target genes (sFRP1 and Smad4) were directly regulated by miR-1260b. The expression of sFRP1 and Smad4 was significantly decreased in prostate cancer tissues. Genistein also increased expression of these two genes via DNA demethylation and histone modifications.
Conclusions:
Our data suggest that genistein exerts its anti-tumour effect via downregulation of miR-1260b that targeted sRRP1 and Smad4 genes in prostate cancer cells. The expression of sFRP1 and Smad4 was also modulated by genistein via DNA methylation or histone modifications in PC cell lines.
Similar content being viewed by others
Main
Prostate cancer (PC) is the second most diagnosed cancer and the sixth leading cause of death in males worldwide (Jemal et al, 2011). In US males however, prostate cancer is the most frequently diagnosed and the second leading cause of death (Siegel et al, 2012). Racial and ethnic disparities exist in prostate cancer incidence and mortality rates globally (Jemal et al, 2011) and generally, the incidence rate of prostate cancer is low in Asia compared with Western countries (Namiki et al, 2010; Gomez et al, 2013; Katanoda et al, 2013; Liu et al, 2013). As the etiology of PC is largely unknown, lower incidence and mortality rates of prostate cancer have been thought to be due to high consumption of soybeans and its products in Asian countries (Shukla and Gupta, 2005; Chan et al, 2006; Jian, 2009; Namiki et al, 2010). In addition, environmental factors may also have an important role in the etiology of prostate cancer (Roemeling et al, 2006). Of the nutritional compounds, genistein is a common dietary isoflavone found in a numerous plants including lupin, fava beans, soybeans and coffee (Kaufman et al, 1997). Genistein has several crucial health benefits such as decreasing blood cholesterol, lowering the risk of cardiovascular diseases and attenuation of postmenopausal problems (Cimino et al, 2012). Previous reports have shown its chemo-preventive properties and anti-cancer effects including anti-invasion, anti-proliferation, pro-apoptotic and modulation of the cell cycle (inhibition of tyrosine kinase, NF-kB, Akt signalling pathways) in prostate cancer cell lines (Xu et al, 2009; Ozten-Kandaş and Bosland, 2011; Ahmad et al, 2013). In in vivo studies, genistein inhibited cell growth and metastasis in a prostate cancer mouse model (Mentor-Marcel et al, 2001; Wang et al, 2004). Based on recent literature, interventional randomized clinical trials have been conducted to examine the effect of genistein on serum and tissue biomarkers in patients with localised prostate cancer where genistein reduced PSA levels (Lazarevic et al, 2011). With regards to its gene regulatory mechanisms, genistein has been reported to upregulate mRNA expression in some tumour suppressor genes including GSTP1 and EPH2 through DNA demethylation of these genes (Vardi et al, 2010). Genistein also produced substantial changes in histone H3K9 acetylation in the promoter region of Wnt antagonist genes (Phillip et al, 2012).
MicroRNAs have emerged as new gene regulators as miRNAs bind to the 3′UTR of target mRNA and repress translation of mRNA to protein or induce mRNA cleavage (Inui et al, 2010). Genistein has been reported to alter expression of some miRNAs in prostate cancer cells (Rabiau et al, 2011; Chiyomaru et al, 2013).
In the present study, we initially looked at the effect of genistein (cell proliferation, invasion, migration, apoptosis, cell cycle, TCF reporter activity) on prostate cancer cell lines. Next we selected microRNA-1260b as a potential candidate oncogenic miRNA whose expression was downregulated by genistein based on microRNA microarray data. We hypothesise that miRNA-1260b may function as an oncogene and is regulated by genistein. We also hypothesise that the target genes of miR-1260b may be regulated by genistein. To test these hypotheses, we performed real-time PCR to determine miR-1260b expression in prostate cancer and benign prostate hyperplasia (BPH) tissues. We also used normal prostate and prostate cancer cell lines (RWPE-1, PC-3, DU-145) and performed in vitro functional analysis. As microRNA exerts its function through regulation of target genes, we identified two target genes based on computer algorithms and performed 3′UTR luciferase assay and Western analyses to examine the direct effect between miR-1260b and its target genes. As expression of the two target genes was increased by genistein, we also investigated the effect of genistein on DNA demethylation and histone modifications of the two target genes in prostate cancer cell lines.
Materials and methods
Ethics statement
Formalin-fixed, paraffin-embedded (FFPE) prostate cancer samples were obtained from the San Francisco Veterans Affairs Medical Center. Written informed consent was obtained from all patients and the study was approved by the UCSF Committee on Human Research (Approval number: H9058-35751-01).
Clinical samples
A total of 30 patients with pathologically confirmed prostate cancer (n=19) and benign prostate hyperplasia (n=11) were enrolled in this study (San Francisco Veterans Affairs Medical Center). Detailed patient information is shown in Supplementary Table S1.
Genistein and transfection
Genistein was purchased from (Sigma-Aldrich, St Louis, MO, USA) and used at a concentration of 25 μ M based on our laboratory’s previous report (Hirata et al, 2013). Prostate cancer cells were treated with DMSO or genistein (25 μ M) for 4 days and used in experiments. Cell culture media was replaced every day.
5-Aza-2′-deoxycytidine treatment
Prostate cancer cell lines were treated with the demethylating agent 5-Aza-2′-deoxycytidine (5Aza; 5 μ M) alone for 4 days or treated with DMSO 4 days.
Cell culture
Normal prostate epithelial cells (RWPE-1; ATCC number-CRL-11609) and prostate cancer cell lines (PC-3; ATCC number-CRL-1435, DU-145; ATCC number-HTB-81) were purchased from the American Type Culture Collection (Manassas, VA, USA). The prostate cancer cell lines were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS). RWPE-1 cells were cultured in keratinocyte-SFM (GIBCO/Invitrogen, Carlsbad, CA, USA). When purchased, permanent stocks of cells were prepared and all cells were stored at −80 degrees until use. Cells were used for experiments within 6 months.
Total RNA, DNA and protein extraction
RNA (microRNA and total RNA) was extracted from FFPE human prostate cancer (n=19) and BPH tissues (n=11) using a miRNeasy FFPE kit (QIAGEN, Valencia, CA, USA) after laser capture microdissection based on a pathologist reviews. RNA (microRNA and total RNA) was also extracted from human cell lines using a miRNeasy mini kit (QIAGEN). DNA was digested with a QIAGEN RNase-Free DNase kit. Genomic DNA was extracted from cell lines using a QIAamp DNA mini kit (QIAGEN). Cells were lysed with RIPA buffer (Pierce, Brebieres, France) containing protease inhibitors (Sigma-Aldrich). Protein quantification was done using a BCA protein assay kit (Pierce).
MicroRNA Microarray
In order to select potential candidate onco-microRNA whose expression was downregulated by genistein, we used commercial microRNA microarray service shown in previously from our labs (Chiyomaru et al, 2013).
Bisulphite DNA sequencing
Genomic DNA was modified with sodium bisulphite using a CpGenome Fast DNA modification kit (CHEMICON International, Temecula, CA, USA). The genomic region and primers used for bisulphite genomic DNA sequencing were shown in Supplementary Figure S1 and Supporting Information Supplementary Table S2. Each PCR reaction was carried out in a total volume of 10 μl consisting of 0.3 μl of each primer (10 mmol l−1, sense and anti-sense, respectively), 1.0 μl of 10 × ExTaq buffer, 0.8 μl of dNTP mixture (2.5 mM each), 0.6 μl of DMSO, 0.1 μl of TaKaRa ExTaq (5 units per μl) (Clontech Laboratories, Mountain View, CA, USA), 1 μl of bisulphite genomic DNA (30 ng μl−1) and 5.9 μl of sterile deionized H2O using a DNA Engine Peltier Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA). PCR conditions were as follows: 5 min at 95 °C; 30–35 cycles of 30 s at 94 °C, 30 s at 56–62 °C, 30 s at 72 °C; and then 5 min at 72 °C. Subsequently, the 3 μl of PCR products were analysed in a 1.5% agarose gel containing ethidium bromide. Then the amplification products were sequenced directly by an outside vendor (TACGen, Richmond, CA, USA).
ChIP assay
ChIP assays were performed on cell line DNA using an Imprint Chromatin Immunoprecipitation Kit (Sigma-Aldrich). Antibody for dimethyl-Histone H3 (Lys9) (H3K9-me2) (catalogue# ab1220, Abcam, Cambridge, MA, USA), trimethyl-histone H3 (Lys9) (H3K9-me3) (catalogue#07-442, EMD Millipore, Billerica, MA, USA), trimethyl-histone H3 (Lys27) (H3K27-me3) (catalogue#07-449, EMD Millipore) was obtained from vendors. The immunoprecipitated DNA was analysed using quantitative real-time PCR with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Quantitative PCR was performed in triplicate with an Applied Biosystems Prism 7500 Fast Sequence Detection System (Applied Biosystems) using the following conditions: 10 min, 95 °C; followed by 40 cycles of 30 s; 95 °C, 60 s; 60 °C. Δ–Δ cycle threshold (Ct) method was used to calculate real-time PCR results. The fold changes related to input ΔCt were calculated as 2−(Δ–ΔC)t. The primer pairs used for ChIP assays are shown in Supporting Information Supplementary Table S2.
MicroRNA and microRNA inhibitor transfection
Anti-miR miRNA inhibitor (negative control (miR-NC inhibitor) or miR-1260b inhibitor (miR-1260b inhibitor), Ambion, Foster City, CA, USA) were transiently transfected into cancer cells with siPORT NeoFX transfection agent (Ambion) according to the manufacturer’s instructions. Pre-miR miRNA precursors (negative control (miR-NC) or hsa-miR-1260b (miR-1260b), Ambion) were transfected into cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
TCF/LEF reporter assay (TOPflash luciferase assay)
TOPflash luciferase assays were performed to assess the effect of genistein on the Wnt signalling pathway. After DMSO (4 days) or genistein (25 μ M-4 days) treatment, cells were trypsinized and re-suspended in 48 well plates overnight. Cells were then transiently co-transfected with TOPflash (Upstate, Lake Placid, NY, USA) and pRL-TK Vector (Promega, Madison, WI, USA) encoding renilla luciferase as an internal control for transfection efficiency using lipofectamine LTX (Invitrogen). To look at the effect of miR-1260b on the beta-catenin dependent pathway, we knocked down miR-1260b with miR-1260b inhibitor (48 h transfection). Cells were trypsinized and re-suspended in 48 well plates overnight and then were transiently co-transfected with TOPflash (Upstate) and pRL-TK Vector (Promega).
Cell viability, cell invasion and wound healing assay
Cell viability was measured 3 days after transfection (miR-NC inhibitor/miR-1260b inhibitor transfectant) with MTS (CellTiter 96 Aqueous One Solution Cell Proliferation Assay, Promega). Cell invasion assays were performed with the CytoSelect 24-well cell invasion assay kit (Cell BioLab, San Diego, CA, USA) according to the manufacturer’s instructions. Transfected cells (miR-NC inhibitor/miR-1260b inhibitor transfectant 48 hours) were re-suspended in culture medium without FBS and placed in the upper chamber in triplicate. After 48 h incubation at 37 °C (5% CO2), cells migrating through the membrane were stained. The results were expressed as invaded cells quantified at OD 560 nm. Wound healing assay was performed with the CytoSelect 24-well wound healing assay kit according to the manufacturer’s instructions. Wound closure was monitored and the percent closure was measured. (Percent closure rate (%)=migrated cell surface area/total surface area × 100)). Data are the mean±s.d. of three independent experiments.
Apoptosis and cell cycle analysis
The apoptotic distribution of the cells in each sample was determined using a FACS (Cell Lab QUANTA SC; Beckman Coulter, Fullerton, CA, USA). The various phases of cells were determined using a DNA stain (4′, 6-diamidino-2-phenylindole). Cell populations (G0/G1, S and G2/M) were differentiated according to 4′, 6-diamidino-2-phenylindole intensity and side scatter-measured cell volume and quantified with Cell Lab QUANTA software. Data are the mean±s.d. of three independent experiments.
Quantitative real-time RT–PCR
Quantitative real-time RT–PCR was performed in triplicate with an Applied Biosystems Prism 7500 Fast Sequence Detection System using TaqMan universal PCR master mix according to the manufacture’s protocol (Applied Biosystems). The TaqMan probes and primers were purchased from Applied Biosystems. RNU48 and GAPDH were used as endogenous control. Levels of RNA expression were determined using the 7500 Fast System SDS software version 1.3.1 (Applied Biosystems). The mRNA and miRNA expression levels were determined using the 2–ΔCt or 2–ΔΔCt method.
Western analysis
Total cell protein (15–20 μg) was used for western blotting. Samples were resolved in 4–20% Precise Protein Gels (Pierce) and transferred to PVDF membranes (Amersham Biosciences, Fairfield, CT, USA). The membranes were immersed in 0.3% skim milk in TBS containing 0.1% Tween 20 for 1 h and probed with primary polyclonal and monoclonal antibody against sFRP1 (#3534, Cell Signaling Technology, Beverly, MA, USA), Smad4 (#9515, Cell Signaling Technology), and beta-tubulin (#2128, Cell Signaling Technology) overnight at 4 °C. Blots were washed in TBS containing 0.1% Tween 20 and labelled with horseradish peroxidase-conjugated secondary anti-mouse or anti-rabbit antibody (Cell Signaling Technology). Proteins were enhanced by chemiluminescence (Amersham ECL plus western blotting detection system) for visualisation. The protein levels were expressed relative to beta-tubulin levels.
Plasmid construction and 3′UTR Luciferase assay
We constructed individual plasmids for each binding site in the 3′UTR of mRNA from potential target genes based on microRNA.org information (Supplementary Figure S2). Then we confirmed miR-1260b binding to the 3′UTR of target gene mRNA by luciferase assay with miR-1260b precursor. PmirGLO Dual-Luciferase miRNA Target Expression Vector was used to perform 3′UTR luciferase assay (Promega). The primer sequences used for plasmid inserts are shown in Supplementary Table S2. For miR-1260b precursor transfection, prostate cancer cells were co-transfected with miR-NC and pmirGLO or miR-1260b and pmirGLO Dual-Luciferase miRNA Target Expression Vectors using Lipofectamine 2000 (Invitrogen). Luciferase activity was assessed using the Dual-Luciferase Reporter Assay System (Promega) (48 h after transfection).
Statistical analysis
All statistical analyses were performed using StatView (version 5; SAS Institute, Cary, NC, USA). Error bars in all the figures represent s.d. Statistical significance was determined using the Student’s t-test or analysis of variance for functional analysis. A P-value of⩽0.05 was regarded as statistically significant.
Results
Effect of genistein on prostate cancer cells
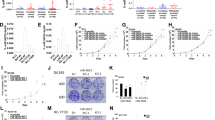
Cancer cell proliferation (Figure 1A), invasion (Figure 1B) and migration (Figure 1C) were significantly decreased after 4 days of genistein treatment (25 μ M) in prostate cancer cell lines (PC-3, DU-145). The percent of apoptotic cells was significantly higher in genistein-treated prostate cancer cells (Figure 1D). While cells in the Go/G1 phase were significantly decreased, in contrast, cells in the G2M phase were significantly increased by genistein treatment (Figure 1E). TCF reporter activity was significantly decreased with genistein in the two prostate cancer cell lines (Figure 1F).
Effect of genistein on prostate cancer cell lines (PC-3 and DU-145). Two cell lines were treated with DMSO (control) or genistein (25μ M) for 4 days. (A) Cell viability assay, (B) invasion assay, (C) wound healing assay, (D) apoptosis assay-FACS based, (E) cell cycle analysis, (F) TOPflash luciferase assay. Error bars represent±s.d.
miRNA-1260b expression is significantly increased in prostate cancer cell lines and significantly decreased by genistein
Based on microarray data (Supplementary Table S3), we have selected miRNA-1260b as a potential onco-miRNA whose expression was downregulated by genistein for further experiments in this study. Initially, we validated microarray data and miR-1260b expression was significantly higher in prostate cancer cell lines (PC-3 and DU-145) compared with the normal prostate cell line (RWPE-1) (Figure 2A). As shown in Figure 2B, the expression of miR-1260b was validated to be significantly decreased by genistein treatment in the two prostate cancer cell lines.
miRNA-1260b expression is significantly increased in prostate cancer tissues
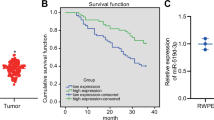
MiR-1260b expression levels in clinical samples (PC-19 samples, BPH-11 samples) were measured by real-time PCR. As shown in Figure 2C, miR-1260b expression was significantly higher in prostate cancer tissues (n=19) compared with BPH tissues (n=11).
The 19 prostate cancer patients were divided into two categories based on the median 2−(ΔCt) in prostate cancer tissues (group cut off point: 2−ΔCt=5.004) and used to associate of miR-1260b expression with overall survival after prostatectomy. The overall survival was shorter in the higher miR-1260b group but was not statistically significant (data not shown).
Effect of microRNA-1260b overexpression on cell viability and migration in a normal prostate cell line
At 48 h after transfection of miR-NC or miR-1260b precursor into RWPE-1 cells, the miR-1260b expression level was verified by real-time PCR (fold change; 967, Figure 3A). Then cell viability (MTS assay) and wound healing assays were performed. We observed significantly increased cell proliferation (Figure 3B) and migration (Figure 3C) in miRNA-1260b transfected cells compared with miR-NC transfected cells.
Effect of microRNA-1260b knockdown on cell viability, invasion and migration in PC cell lines
After transfection (48 h) of inhibitor-NC or miR-1260b inhibitor into PC-3 and DU-145 cells, miR-1260b knockdown was verified by real-time RT–PCR (0.031, 0.026, respectively) (Figure 4A). We observed a significant decrease in cell proliferation (Figure 4B), invasion (Figure 4C) and migration (Figure 4D) in miRNA-1260b knockdown PC-3 and DU-145 cells compared with inhibitor-NC transfected cells. The percent of apoptotic cells was significantly increased by miR-1260b knockdown (Figure 4E). Also, the percent of Go-G1 phase cells was significantly decreased while the percent of G2M phase cells was significantly increased in miR-1260b inhibitor transfected prostate cancer cells (Figure 4F). TOPflash luciferase activity was significantly decreased by miR-1260b knockdown (Figure 4G).
Effect of miR-1260b knockdown on prostate cancer cells (PC-3, DU-145). Two prostate cancer cell lines (PC-3 and DU-145) were transiently transfected with either miR-1260b inhibitor or miR-negative control (miR-NC inhibitor). (A) Relative miR-1260b expression (miR-NC inhibitor or miR-1260b inhibitor transfected PC cells), (B) cell viability assay (miR-NC inhibitor or miR-1260b inhibitor transfected PC cells), (C) invasion assay, (D) wound healing assay (16–17 h). (E) Apoptosis assay, (F) cell cycle analysis, (G) TCF reporter assay. Error bars represent ±s.d.
Expression of sFRP1 and Smad4 in PC and BPH tissues
We identified two tumour suppressor genes (sFRP1, Smad4) as potential target genes for miR-1260b based on computer algorithms (microRNA.org), and we compared the expression of these two genes in PC and BPH tissues. As shown in Figure 5A, gene expression was significantly lower in PC compared with BPH tissues. In addition, expression of both target genes (sFRP1 and Smad4) was significantly lower in prostate cancer cell lines (PC-3 and DU-145) compared with the normal prostate cell line (RWPE-1) (Figure 5B).
Expression of miR-1260b target genes ( sFRP1 and Smad4 ) in BPH and PC tissues and relationship with miR-1260b in prostate cancer cell lines. (A) sFRP1 expression level in BPH and PC tissues, (B) Smad4 expression level in BPH and PC tissues, (C) 3′UTR Luciferase assay (miR-NC and miR-1260b precursor), (D) expression of sFRP1, Smad4 and beta-tubulin protein in miR-NC inhibitor or miR-1260b inhibitor transfected prostate cancer cells (PC-3, DU-145). Error bars represent±s.d.
miR-1260b directly regulates sFRP1 and Smad4
The relative 3′UTR luciferase activity is shown as a ratio of firefly and renilla luciferase activity for each sample. As shown in Figure 5C, the relative luciferase activity was significantly decreased at position 2500 (sFRP1) and at position 2914 (Smad4) in miR-1260b precursor transfected cells (Figure 5C). As target gene protein expression is low in PC-3 and DU-145, we performed western analysis to observe any changes in protein expression with miR-1260b inhibitor. As shown in Figure 5D, protein expression of target genes was significantly increased after miR-1260b inhibitor transfection. This result suggests that the sFRP1 and Smad4 mRNAs are direct targets of miR-1260b.
5-Aza effect on sFRP1 and Smad4 gene expression in prostate cancer cell lines
Expression of sFRP1 and Smad4 were significantly increased by 5Aza treatment in PC-3 and DU-145 cells (Supplementary Figure S3).
Genistein effect on DNA methylation in target genes of miR-1260b
Expression of sFRP1 and Smad4 were significantly increased by genistein treatment in PC-3 and DU-145 cells (Figure 6A). As there are CpG islands in the promoter region of the sFRP1 and Smad4 genes based on a CpG island prediction software (Li and Dahiya, 2002), we looked at these CpG islands to determine the demethylation effect of genistein in prostate cancer cell line (PC-3). As shown in Figure 6B, CpG islands are completely methylated in the promoter region of the sFRP1 gene by sodium bisulphite treatment and 2 of 15 CpG sites in the region were changed to TG by genistein treatment (Figure 6B). However, CpG sites in the promoter region of the Smad4 gene were not methylated in control cells (Figure 6C).
Effect of genistein on sFRP1 and Smad4 genes in prostate cancer cells. (A) sFRP1 and Smad4 expression in DMSO (control) and genistein 25 μ M (G25) treated prostate cancer cells. (B) Representative bisulphite modified genomic DNA sequencing of the sFRP1 promoter region from control and genistein-treated PC-3 cells. (C) Representative bisulphite modified genomic DNA sequencing of the Smad4 promoter region from control and genistein-treated PC-3 cells. (D) Quantification of ChIP real-time PCR analysis in PC-3 cells in sFRP1 and Smad4 genes promoter region in control and genistein-treated 786-O cells.
Effect of genistein on histone modification in target genes of miR-1260b
We also used ChIP assay to determine the effect of genistein on histone modification in the sFRP1 and Smad4 genes. The histone associated DNAs that were immunoprecipitated with antibody against H3K9-me2, H3K9-me3 and H3K27-me3 were individually amplified with primer sets covering the gene promoter regions. The levels of H3K9-me2, H3K9-me3 and H3K27-me3 were significantly decreased by genistein compared with control cells in both genes (Figure 6D).
Discussion
The chemo prevention effect of genistein on prostate cancer has been investigated both in vitro and in vivo as interventional clinical trials for prostate cancer treatment. Based on previous reports, genistein inhibits MDM2, Akt and NF-kappaB activity, resulting in inhibition of cell growth and induction of apoptosis in prostate cancer cells (Li and Sarkar, 2002; Li et al, 2005). In animal models, genistein has been reported to decrease prostate tumour volume (Mentor-Marcel et al, 2001; Fritz et al, 2002). Many clinical trials (phases I and phases II) using genistein for treatment of prostate cancer patients have been carried out (Lazarevic et al, 2011) and clinical trials are on going to justify the usefulness of genistein for prostate cancer prevention and therapy.
In this study, we initially investigated the effect of genistein on prostate cancer cell lines and found that genistein decreased cell proliferation, migration, invasion, TCF reporter activity and increased apoptosis in these cells. Our results are similar to those of previous reports (Ahmad et al, 2013; Hirata et al, 2013). The main aim of this study is to determine the effect of genistein and miRNA in prostate cancer cell lines and miRNA target genes. Thus, after examining the effect of genistein on prostate cancer cells, we selected miRNA-1260b, whose expression was significantly downregulated by genistein based on microRNA microarray data, suggesting that miR-1260b is an oncogenic miRNA. We looked at miRNA-1260b expression in human prostate cancer and BPH tissues and found that miR-1260b expression was significantly higher in prostate cancer compared with BPH samples. To study the functional mechanism of miR-1260b in vitro, we used two procedures as follows: (1) overexpression of miR-1260b in a normal prostate cell line (RWPE-1), (2) knockdown of miR-1260b in prostate cancer cell lines (PC-3 and DU-145). After overexpressing miR-1260b in RWPE-1 cells, cell proliferation and wound healing (cell migration ability) were significantly increased. After miR-1260b knockdown in prostate cancer cells (PC-3 and DU-145), cell proliferation, migration and invasion were decreased while apoptosis was significantly promoted in prostate cancer cell lines. At present, no other miR-1260 studies have been carried out except for our laboratory’s study on RCC (Hirata et al, 2013). However, considering both studies together, the miR-1260b expression data in clinical samples and our in vitro studies indicate that miR-1260b may be an oncogenic miRNA.
As miRNAs exert their function by regulating target genes, we identified two target genes of miR-1260b based on computer algorithms (microRNA.org). Two potential target genes (sFRP1 and Smad4) were identified and 3′UTR luciferase assay and western analyses showed that sFRP1 and Smad4 expression was regulated by miR-1260b. Expression of both sFRP1 and Smad4 was significantly lower in prostate cancer tissues and expression of these two tumour suppressor genes is significantly increased by genistein.
Wnt antagonists include the secreted frizzled-related protein (sFRP) family, Dickkopf family, Wnt inhibitory factor 1 and Cerberus. Several members of the sFRP family downregulate Wnt signalling in cancer cells (Kawano and Kypta, 2003). sFRP1 has been reported to be downregulated epigenetically and overexpression of sFRP1 inhibited cell growth in several cancers (colon, breast, oesophagus, brain), indicating that loss of sFRP1 contributes to activation of Wnt signalling in several cancers (Suzuki et al, 2004, 2008; Clément et al, 2006; Götze et al, 2010). The sFRP1 has also been reported to be epigenetically silenced in prostate cancer cell lines (PC-3 and DU-145) and prostate cancer tissues compared with normal prostate tissues (Lodygin et al, 2005). In this study, we looked for potential demethylation effects of genistein on the sFRP1 gene promoter in prostate cancer cell lines. Bisulphite genomic sequencing showed that the percentage of CpG sites demethylated by genistein was 13% (2/15 CpG sites) in the promoter region of the sFRP1 gene. With DU-145 cells, no CpG sites were altered by genistein. Therefore, the demethylation effect of genistein may only partially contribute to sFRP1 gene regulation. Phillip et al (2012) investigated demethylation of the sFRP1 gene by genistein (20 μ M, 6 days) but no effect was found in prostate cancer cell lines (PC-3 and DU-145) and our results are very similar to theirs. Next, we focused on three histones such as H3K9-me2, H3K9-me3 and H3K27-me3, which are thought to repress gene expression (Swigut and Wysocka, 2007: Wang et al, 2008). Interestingly genistein caused a significant reduction of these repressive histones at three different positions in the gene promoter. Thus, the regulation of sFRP1 by genistein may be mainly by chromatin histone modifications rather than DNA methylation.
The other target gene Smad4 has been extensively studied as a tumour suppressor gene in many cancers. Smad4 was first identified as tumour suppressor gene in pancreatic cancer in 1996 (Hahn et al, 1996). The Smad4 gene is located on chromosome 18q21.1 and is a member of the Smad family, which are mediators of the transforming growth factor beta signalling pathway (Riggins et al, 1997). Deficiency of Smad4 initiates or promotes the development and progression of many cancers including pancreas, gastroenterological and breast cancers (Giehl et al, 2007; Yang and Yang, 2010). Loss of Smad4 expression has also been related to the progression of prostate cancer (Horvath et al, 2004). Other studies indicate that Smad4 is downregulated in metastatic prostate cancer tissues and knockdown of the Smad4 gene showed significantly enhanced metastatic potential to lung with PC-3 cells implanted in the renal capsule of an immunocompromised nude mouse (Ding et al, 2011). Yin et al, (2001) investigated the loss of heterozygosity on chromosome 18q21 in prostate cancer tissues but no alleles were lost, suggesting that Smad4 is not inactivated by allelic loss in prostate cancer. According to CpG prediction software, there are large CpG islands in the promoter region of the Smad4 gene and the Smad4 expression was increased by demethylation agent, 5-aza-dC treatment in two prostate cancer cell lines (Supplementary Figure S3). Thus, while we initially expected to observe hypermethylation in the CpG islands of the Smad4 gene in prostate cancer cell lines, no evidence of Smad4 gene methylation was found in PC-3 and DU-145 cells. Our results are very similar to previous DNA methylation result in PC-3 and DU-145 cells (Perry et al, 2007).
According to previous papers, 5-aza-dC restored RASSF1A expression through a loss of gene-silencing H3K9m2 and H3K9-me3 methylation in the promoter region (Lambrot and Kimmins, 2011). In order to look at Smad4 regulation by genistein, we performed ChIP assays similar to that for sFRP1 and found that some histones were reduced by genistein in PC-3 cells at the promoter region. Although there was no methylation in the DNA promoter region of the Smad4 gene, the regulatory effect of genistein on Smad4 was by histone modifications and miRNA. In this regard, Øster et al, (2013) has shown that the expression of SRPX2 gene (oncogene) was increased by hypomethylation of DNA CpG islands, and also by downregulation miR149, which targets SRPX2, resulting in increased SRPX2 gene expression in colon cancer. In normal colon cells, expression of SRPX2 was decreased by DNA methylation and miR149 caused degradation of SRPX2 mRNA. Thus, it was concluded that SRPX2 expression was regulated through DNA methylation and miRNA-149 in colon cancer. Similarly, our results suggested that sFRP1 and Smad4 expression may be downregulated synergistically by miR-1260b and by DNA methylation/histone modification, not exclusively one or the other. Indeed, as shown in Figures 5 and 6, Supplementary Figure S3, the sFRP1 and Smad4 expression level was significantly increased both by 5Az treatment (Chip assay) and by miR-1260b inhibitor transfection (luciferase assay).
As we investigated only 19 clinical samples, additional studies will be needed to elucidate the role of miR-1260b in prostate cancer and its use in clinical applications.
In conclusion, this is the first report documenting that genistein exerts its anti-tumour effect via downregulation of miR-1260b that targets sFRP1 and Smad4 in prostate cancer cells. Knockdown of miR-1260b by a specific inhibitor enhanced expression of tumour suppressor genes sFRP1 and Smad4 in prostate cancer cells, suggesting potential therapeutic benefit for prostate cancer treatment. In addition, the expression of sFRP1 and Smad4 was also modulated by genistein via DNA methylation or histone modifications in prostate cancer cell lines, indicating the potential importance of diet in regulating these two tumour suppressor genes.
Change history
18 March 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
28 July 2023
An Editorial Expression of Concern to this paper has been published: https://doi.org/10.1038/s41416-023-02365-0
References
Ahmad A, Biersack B, Li Y, Bao B, Kong D, Ali S, Banerjee S, Sarkar FH (2013) Perspectives on the role of isoflavones in prostate cancer. AAPS J 15: 991–1000.
Chan JM, Holick CN, Leitzmann MF, Rimm EB, Willett WC, Stampfer MJ, Giovannucci EL (2006) Diet after diagnosis and the risk of prostate cancer progression, recurrence, and death (United States). Cancer Causes Control 17: 199–208.
Chiyomaru T, Yamamura S, Fukuhara S, Hidaka H, Majid S, Saini S, Arora S, Deng G, Shahryari V, Chang I, Tanaka Y, Tabatabai ZL, Enokida H, Seki N, Nakagawa M, Dahiya R (2013) Genistein up-regulates tumor suppressor microRNA-574-3p in prostate cancer. PLoS One 8: e58929.
Cimino S, Sortino G, Favilla V, Castelli T, Madonia M, Sansalone S, Russo GI, Morgia G (2012) Polyphenols: key issues involved in chemoprevention of prostate cancer. Oxid Med Cell Longev 2012: 632959.
Clément G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J (2006) Alterations of the Wnt signaling pathway during the neoplastic progression of Barrett’s esophagus. Oncogene 25: 3084–3092.
Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, Hoshida Y, Hiller D, Hu B, Jiang S, Zheng H, Stegh AH, Scott KL, Signoretti S, Bardeesy N, Wang YA, Hill DE, Golub TR, Stampfer MJ, Wong WH, Loda M, Mucci L, Chin L, DePinho RA (2011) SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470: 269–273.
Fritz WA, Eltoum IE, Cotroneo MS, Lamartiniere CA (2002) Genistein alters growth but is not toxic to the rat prostate. J Nutr 132: 3007–3011.
Giehl K, Imamichi Y, Menke A (2007) Smad4-independent TGF-beta signaling in tumor cell migration. Cells Tissues Organs 185: 123–130.
Gomez SL, Noone AM, Lichtensztajn DY, Scoppa S, Gibson JT, Liu L, Morris C, Kwong S, Fish K, Wilkens LR, Goodman MT, Deapen D, Miller BA (2013) Cancer incidence trends among asian american populations in the United States, 1990-2008. J Natl Cancer Inst 105: 1096–1110.
Götze S, Wolter M, Reifenberger G, Müller O, Sievers S (2010) Frequent promoter hypermethylation of Wnt pathway inhibitor genes in malignant astrocytic gliomas. Int J Cancer 126: 2584–2593.
Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE (1996) DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271: 350–353.
Hirata H, Ueno K, Nakajima K, Tabatabai ZL, Hinoda Y, Ishii N, Dahiya R (2013) Genistein downregulates onco-miR-1260b and inhibits Wnt-signalling in renal cancer cells. Br J Cancer 108: 2070–2078.
Horvath LG, Henshall SM, Kench JG, Turner JJ, Golovsky D, Brenner PC, O'Neill GF, Kooner R, Stricker PD, Grygiel JJ, Sutherland RL (2004) Loss of BMP2, Smad8, and Smad4 expression in prostate cancer progression. Prostate 59: 234–242.
Inui M, Martello G, Piccolo S (2010) MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11: 252–263.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90.
Jian L (2009) Soy, isoflavones, and prostate cancer. Mol Nutr Food Res 53: 217–226.
Katanoda K, Matsuda T, Matsuda A, Shibata A, Nishino Y, Fujita M, Soda M, Ioka A, Sobue T, Nishimoto H (2013) An updated report of the trends in cancer incidence and mortality in Japan. Jpn J Clin Oncol 43: 492–507.
Kaufman PB, Duke JA, Brielmann H, Boik J, Hoyt JE (1997) A comparative survey of leguminous plants as sources of the isoflavones, genistein and daidzein: implications for human nutrition and health. J Altern Complement Med 3: 7–12.
Kawano Y, Kypta R (2003) Secreted antagonists of the Wnt signalling pathway. J Cell Sci 116: 2627–2634.
Lambrot R, Kimmins S (2011) Histone methylation is a critical regulator of the abnormal expression of POU5F1 and RASSF1A in testis cancer cell lines. Int J Androl 34: 110–123.
Lazarevic B, Boezelijn G, Diep LM, Kvernrod K, Ogren O, Ramberg H, Moen A, Wessel N, Berg RE, Egge-Jacobsen W, Hammarstrom C, Svindland A, Kucuk O, Saatcioglu F, Taskèn KA, Karlsen SJ (2011) Efficacy and safety of short-term genistein intervention in patients with localized prostate cancer prior to radical prostatectomy: a randomized, placebo-controlled, double-blind Phase 2 clinical trial. Nutr Cancer 63: 889–898.
Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18: 1427–1431.
Li M, Zhang Z, Hill DL, Chen X, Wang H, Zhang R (2005) Genistein, a dietary isoflavone, down-regulates the MDM2 oncogene at both transcriptional and posttranslational levels. Cancer Res 65: 8200–8208.
Li Y, Sarkar FH (2002) Inhibition of nuclear factor kappaB activation in PC3 cells by genistein is mediated via Akt signaling pathway. Clin Cancer Res 8: 2369–2377.
Liu L, Noone AM, Gomez SL, Scoppa S, Gibson JT, Lichtensztajn D, Fish K, Wilkens LR, Goodman MT, Morris C, Kwong S, Deapen D, Miller BA (2013) Cancer incidence trends among native Hawaiians and other Pacific islanders in the United States, 1990-2008. J Natl Cancer Inst 105: 1086–1095.
Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H (2005) Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res 65: 4218–4227.
Mentor-Marcel R, Lamartiniere CA, Eltoum IE, Greenberg NM, Elgavish A (2001) Genistein in the diet reduces the incidence of poorly differentiated prostatic adenocarcinoma in transgenic mice (TRAMP). Cancer Res 61: 6777–6782.
Namiki M, Akaza H, Lee SE, Song JM, Umbas R, Zhou L, Lee BC, Cheng C, Chung MK, Fukagai T, Hinotsu S, Horie S (2010) Prostate cancer working group report. Jpn J Clin Oncol Suppl 1: i70–i75.
Øster B, Linnet L, Christensen LL, Thorsen K, Ongen H, Dermitzakis ET, Sandoval J, Moran S, Esteller M, Hansen TF, Lamy P COLOFOL steering group Laurberg S, Ørntoft TF, Andersen CL (2013) Non-CpG island promoter hypomethylation and miR-149 regulate the expression of SRPX2 in colorectal cancer. Int J Cancer 132: 2303–2315.
Ozten-Kandaş N, Bosland MC (2011) Chemoprevention of prostate cancer: natural compounds, antiandrogens, and antioxidants—in vivo evidence. J Carcinog 10: 27.
Perry AS, Liyanage H, Lawler M, Woodson K (2007) Discovery of DNA hypermethylation using a DHPLC screening strategy. Epigenetics 2: 43–49.
Phillip CJ, Giardina CK, Bilir B, Cutler DJ, Lai YH, Kucuk O, Moreno CS (2012) Genistein cooperates with the histone deacetylase inhibitor vorinostat to induce cell death in prostate cancer cells. BMC Cancer 12: 145.
Rabiau N, Trraf HK, Adjakly M, Bosviel R, Guy L, Fontana L, Bignon YJ, Bernard-Gallon DJ (2011) miRNAs differentially expressed in prostate cancer cell lines after soy treatment. In Vivo 25: 917–921.
Riggins GJ, Kinzler KW, Vogelstein B, Thiagalingam S (1997) Frequency of Smad gene mutations in human cancers. Cancer Res 57: 2578–2580.
Roemeling S, Roobol MJ, de Vries SH, Gosselaar C, van der Kwast TH, Schröder FH (2006) Prevalence, treatment modalities and prognosis of familial prostate cancer in a screened population. J Urol 175: 1332–1336.
Shukla S, Gupta S (2005) Dietary agents in the chemoprevention of prostate cancer. Nutr Cancer 53: 18–32.
Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62: 10–29.
Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, Mori M, Hirata K, Imai K, Shinomura Y, Baylin SB, Tokino T (2008) Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer 98: 1147–1156.
Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, Toyota M, Tokino T, Hinoda Y, Imai K, Herman JG, Baylin SB (2004) Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 36: 417–422.
Swigut T, Wysocka J (2007) H3K27 demethylases, at long last. Cell 131: 29–32.
Vardi A, Bosviel R, Rabiau N, Adjakly M, Satih S, Dechelotte P, Boiteux JP, Fontana L, Bignon YJ, Guy L, Bernard-Gallon DJ (2010) Soy phytoestrogens modify DNA methylation of GSTP1, RASSF1A, EPH2 and BRCA1 promoter in prostate cancer cells. In Vivo 24: 393–400.
Wang J, Eltoum IE, Lamartiniere CA (2004) Genistein alters growth factor signaling in transgenic prostate model (TRAMP). Mol Cell Endocrinol 219: 171–180.
Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903.
Xu L, Ding Y, Catalona WJ, Yang XJ, Anderson WF, Jovanovic B, Wellman K, Killmer J, Huang X, Scheidt KA, Montgomery RB, Bergan RC (2009) MEK4 function, genistein treatment, and invasion of human prostate cancer cells. J Natl Cancer Inst 101: 1141–1155.
Yang G, Yang X (2010) Smad4-mediated TGF-beta signaling in tumorigenesis. Int J Biol Sci 6: 1–8.
Yin Z, Babaian RJ, Troncoso P, Strom SS, Spitz MR, Caudell JJ, Stein JD, Kagan J (2001) Limiting the location of putative human prostate cancer tumor suppressor genes on chromosome 18q. Oncogene 20: 2273–2280.
Acknowledgements
We thank Dr Roger Erickson for his support and assistance with the preparation of the manuscript. This study was supported by National Center for Research Resources of the National Institutes of Health through Grant Number RO1CA138642, RO1CA130860, RO1CA160079, I01BX001123, VA Merit Review, VA Program Project (PI: R Dahiya) and Yamada Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Hirata, H., Hinoda, Y., Shahryari, V. et al. Genistein downregulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. Br J Cancer 110, 1645–1654 (2014). https://doi.org/10.1038/bjc.2014.48
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.48
Keywords
This article is cited by
-
High expression level of miR-1260 family in the peripheral blood of patients with ovarian carcinoma
Journal of Ovarian Research (2021)
-
MicroRNA mediated therapeutic effects of natural agents in prostate cancer
Molecular Biology Reports (2021)
-
Clinical relevance and functional significance of cell-free microRNA-1260b expression profiles in infiltrative myxofibrosarcoma
Scientific Reports (2020)
-
Usefulness of serum microRNA as a predictive marker of recurrence and prognosis in biliary tract cancer after radical surgery
Scientific Reports (2019)
-
miR-1260b promotes cell migration and invasion of hepatocellular carcinoma by targeting the regulator of G-protein signaling 22
Biotechnology Letters (2018)