Abstract
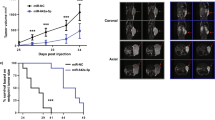
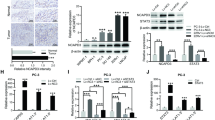
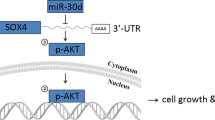
For human prostate cancer, the chromosome 8p21 locus, which contains NKX3.1 and the microRNA (miR)-3622 family (miR-3622a/b), is a frequently deleted region. Thus, miR-3622 is proposed as a suppressor for prostate cancer, but its role remains debatable. In the present study, we found that expression of miR-3622a was lower, whereas expression of miR-3622b-3p was higher in human prostate cancer tissues than in normal prostate tissues. miR-3622a-3p inhibited cell migration and invasion of human prostate cancer cells, whereas miR-3622b-3p facilitated cell proliferation, migration, and invasion. To address the opposing roles of miR-3622 family members in various human prostate cancer cell lines, we knocked out (KO) endogenous miR-3622, including both miR-3622a/b. Our results showed that miR-3622 KO reduced cell proliferation, migration, and invasion in vitro and tumor growth and metastasis in vivo. Functional analyses revealed that miR-3622 regulated the p53-downstream gene network, including AIFM2, c-MYC, and p21, to control apoptosis and the cell cycle. Furthermore, using CRISPR interference, miRNA/mRNA immunoprecipitation assays, and dual-luciferase assays, we established that AIFM2, a direct target of miR-3622b-3p, is responsible for miR-3622 KO-induced apoptosis. We identified an miR-3622-AIFM2 axis that contributes to oncogenic function during tumor progression. In addition, miR-3622 KO inhibited the epithelial–mesenchymal transition involved in prostate cancer metastasis via upregulation of vimentin. The results show that miR-3622b-3p is upregulated in human prostate cancers and has an oncogenic function in tumor progression and metastasis via repression of p53 signaling, especially through an miR-3622-AIFM2 axis. In contrast, for human prostate cancer, deletion of the miR-3622 locus at 8p21 reduced the oncogenic effects on tumor progression and metastasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Rebello RJ, Oing C, Knudsen KE, Loeb S, Johnson DC, Reiter RE, et al. Prostate cancer. Nat Rev Dis Prim. 2021;7:9.
Wallis CJ, Nam RK. Prostate cancer genetics: a review. EJIFCC. 2015;26:79–91.
Schoenborn JR, Nelson P, Fang M. Genomic profiling defines subtypes of prostate cancer with the potential for therapeutic stratification. Clin Cancer Res. 2013;19:4058–66.
Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–6.
Wang Y, Li X, Liu W, Li B, Chen D, Hu F, et al. MicroRNA-1205, encoded on chromosome 8q24, targets EGLN3 to induce cell growth and contributes to risk of castration-resistant prostate cancer. Oncogene. 2019;38:4820–34.
Kagan J, Stein J, Babaian RJ, Joe YS, Pisters LL, Glassman AB, et al. Homozygous deletions at 8p22 and 8p21 in prostate cancer implicate these regions as the sites for candidate tumor suppressor genes. Oncogene. 1995;11:2121–6.
Bucay N, Sekhon K, Majid S, Yamamura S, Shahryari V, Tabatabai ZL, et al. Novel tumor suppressor microRNA at frequently deleted chromosomal region 8p21 regulates epidermal growth factor receptor in prostate cancer. Oncotarget. 2016;7:70388–403.
Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15:565–76.
Fabris L, Ceder Y, Chinnaiyan AM, Jenster GW, Sorensen KD, Tomlins S, et al. The potential of MicroRNAs as prostate cancer biomarkers. Eur Urol. 2016;70:312–22.
Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016;76:3666–70.
Chang S, Sun G, Zhang D, Li Q, Qian H. MiR-3622a-3p acts as a tumor suppressor in colorectal cancer by reducing stemness features and EMT through targeting spalt-like transcription factor 4. Cell Death Dis. 2020;11:592.
Bucay N, Bhagirath D, Sekhon K, Yang T, Fukuhara S, Majid S, et al. A novel microRNA regulator of prostate cancer epithelial-mesenchymal transition. Cell Death Differ. 2017;24:1263–74.
Bhagirath D, Yang TL, Tabatabai ZL, Shahryari V, Majid S, Dahiya R, et al. Role of a novel race-related tumor suppressor microRNA located in frequently deleted chromosomal locus 8p21 in prostate cancer progression. Carcinogenesis. 2019;40:633–42.
Lu M, Wang T, He M, Cheng W, Yan T, Huang Z, et al. Tumor suppressor role of miR-3622b-5p in ERBB2-positive cancer. Oncotarget. 2017;8:23008–19.
Vernon M, Lambert B, Meryet-Figuiere M, Brotin E, Weiswald LB, Paysant H, et al. Functional miRNA screening identifies wide-ranging antitumor properties of miR-3622b-5p and reveals a new therapeutic combination strategy in ovarian tumor organoids. Mol Cancer Ther. 2020;19:1506–19.
Fu S, Luan T, Jiang C, Huang Y, Li N, Wang H, et al. miR-3622a promotes proliferation and invasion of bladder cancer cells by downregulating LASS2. Gene. 2019;701:23–31.
Theodore S, Sharp S, Zhou J, Turner T, Li H, Miki J, et al. Establishment and characterization of a pair of non-malignant and malignant tumor derived cell lines from an African American prostate cancer patient. Int J Oncol. 2010;37:1477–82.
Wu M, Xu LG, Su T, Tian Y, Zhai Z, Shu HB. AMID is a p53-inducible gene downregulated in tumors. Oncogene. 2004;23:6815–9.
Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–23.
Lu J, Chen J, Xu N, Wu J, Kang Y, Shen T, et al. Activation of AIFM2 enhances apoptosis of human lung cancer cells undergoing toxicological stress. Toxicol Lett. 2016;258:227–36.
Tao YF, Xu LX, Lu J, Hu SY, Fang F, Cao L, et al. Early B-cell factor 3 (EBF3) is a novel tumor suppressor gene with promoter hypermethylation in pediatric acute myeloid leukemia. J Exp Clin Cancer Res. 2015;34:4.
Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–92.
Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–8.
el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25.
Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol. 2005;25:7423–31.
Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–5.
Wang M, Zhang G, Zhang Y, Cui X, Wang S, Gao S, et al. Fibrinogen alpha chain knockout promotes tumor growth and metastasis through integrin-AKT signaling pathway in lung cancer. Mol Cancer Res. 2020;18:943–54.
Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–61.
Liu R, Yi B, Wei S, Yang WH, Hart KM, Chauhan P, et al. FOXP3-miR-146-NF-kappaB Axis and therapy for precancerous lesions in prostate. Cancer Res. 2015;75:1714–24.
Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 2009;16:336–46.
Liu R, Liu C, Chen D, Yang WH, Liu X, Liu CG, et al. FOXP3 controls an miR-146/NF-kappaB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. 2015;75:1703–13.
Liu R, Wang L, Chen G, Katoh H, Chen C, Liu Y, et al. FOXP3 up-regulates p21 expression by site-specific inhibition of histone deacetylase 2/histone deacetylase 4 association to the locus. Cancer Res. 2009;69:2252–9.
Zhang W, Yi B, Wang C, Chen D, Bae S, Wei S, et al. Silencing of CD24 enhances the PRIMA-1-induced restoration of mutant p53 in prostate cancer cells. Clin Cancer Res. 2016;22:2545–54.
Wang L, Liu R, Ye P, Wong C, Chen GY, Zhou P, et al. Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun. 2015;6:5909.
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005.
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Acknowledgements
We thank Dr. Donald Hill for editorial assistance in preparing this paper. This work was supported by grants from the National Cancer Institute (CA118948 for LW), the Department of Defense (W81XWH-21-1-0100 for LW and W81XWH-15-1-0323 and W81XWH-20-1-0426 for RL) and the Mike Slive Foundation for Prostate Cancer Research (RL). Results are based, in part, upon data generated by TargetScan (http://www.targetscan.org/), the UALCAN portal (http://ualcan.path.uab.edu/), and the cBioPortal (http://www.cbioportal.org).
Author information
Authors and Affiliations
Contributions
Conception, design, and financial support: RL, LW. Development of methodology:YZ, JZ, LW, and RL. Acquisition of data (bench/animal works, acquired data, etc.): YZ, ZX, WW, ZL, CZ, ML, and FH. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): YZ, SB, RL, and LW. Writing, review, and/or revision of the paper: YZ, RL, and LW. Pathology analysis: YZ, SW, and LW. Study supervision: RL, LW.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, Y., Xu, Z., Wen, W. et al. The microRNA-3622 family at the 8p21 locus exerts oncogenic effects by regulating the p53-downstream gene network in prostate cancer progression. Oncogene 41, 3186–3196 (2022). https://doi.org/10.1038/s41388-022-02289-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02289-8
This article is cited by
-
FSP1-mediated ferroptosis in cancer: from mechanisms to therapeutic applications
Apoptosis (2024)
-
Knockdown of TACC3 inhibits tumor cell proliferation and increases chemosensitivity in pancreatic cancer
Cell Death & Disease (2023)