Abstract
Background:
Developmental arrest of fetal germ cells may lead to neoplastic transformation and formation of germ cell tumours via carcinoma in situ (CIS) cells. Normal fetal germ cell development requires complete erasure and re-establishment of DNA methylation. In contrast to normal spermatogonia, the genome of CIS cells remains unmethylated in the adult testis. We here investigated the possible active and passive pathways that can sustain the CIS genome hypomethylated in the adult testis.
Methods:
The levels of 5-methyl-cytosine (5mC) and 5-hydroxy-methyl-cytosine (5hmC) in DNA from micro-dissected CIS cells were assessed by quantitative measurements. The expression of TET1, TET2, APOBEC1, MBD4, APEX1, PARP1, DNMT1, DNMT3A, DNMT3B and DNMT3L in adult testis specimens with CIS and in human fetal testis was investigated by immunohistochemistry and immunofluorescence.
Results:
DNA from micro-dissected CIS cells contained very low levels of 5hmC produced by ten eleven translocation (TET) enzymes. CIS cells and fetal germ cells expressed the suggested initiator of active demethylation, APOBEC1, and the base excision repair proteins MBD4, APEX1 and PARP1, whereas TETs – the alternative initiators were absent. Both maintenance and de novo methyltransferases were detected in CIS cells.
Conclusion:
The data are consistent with the presence of an active DNA de-methylation pathway in CIS cells. The hypomethylated genome of CIS cells may contribute to phenotypic plasticity and invasive capabilities of this testicular cancer precursor.
Similar content being viewed by others
Main
In most western countries, testicular germ cell cancer (TGCC) is the most common cancer encountered in men aged 15–45 years (Huyghe et al, 2003; Chia et al, 2010). Although TGCC shows a strong familial risk compared to other cancers, recent association studies (Chung et al, 2013; Ruark et al, 2013) have only been able to explain a genetic cause in less than a quarter of cases leaving still a major part of the cases to be caused by unknown gene variants and by environmental factors (Turnbull and Rahman, 2011). The causal influence of environmental factors is substantiated by a rapid increase in incidence of TGCC in recent decades and differences in prevalence between first- and second-generation immigrant populations (Hemminki and Li, 2002).
The pathogenic model for TGCC suggests that the tumours develop from a precursor cell; the carcinoma in situ (CIS) cell (Skakkebaek, 1972). Carcinoma in situ is also described in the literature as intratubular germ cell neoplasia unclassified or testicular intraepithelial neoplasia. The core event in the pathogenesis of CIS is the developmental arrest of primordial germ cells (PGCs) or gonocytes, which remain locked in an immature state as ‘dormant’ or pre-CIS cells during fetal and postnatal life. At puberty, CIS cells proliferate and gain invasive capacity, eventually resulting in the development of a seminoma, a non-seminoma or a combined tumour (Rajpert-De Meyts, 2006).
Morphological and immunohistochemical studies have indicated that CIS cells resemble fetal germ cells (Nielsen et al, 1974; Berthelsen et al, 1979; Albrechtsen et al, 1982; Skakkebaek et al, 1987) and more recent gene expression analyses have provided further evidence for this similarity. In analogy to PGCs and gonocytes, CIS cells express transcription factors associated with embryonic stem cell pluripotency, including POU5F1 (OCT-3/4), NANOG, T1A-2, MYCL1, GDF3, DPPA4, DPPA5, KIT and TFAP2C (AP-2γ) (Looijenga et al, 2003; Sperger et al, 2003; Almstrup et al, 2004; Hoei-Hansen et al, 2004, 2005; Skotheim et al, 2005; Almstrup et al, 2007; Biermann et al, 2007; Sonne et al, 2009a). In addition, recent studies have shown that the overall epigenetic features of CIS cells, including the miRNA profile, shows similarities with the pattern found in fetal germ cells (Almstrup et al, 2010; Novotny et al, 2012). The CIS cells had an open and permissive chromatin structure similar to observations in fetal germ cells (Almstrup et al, 2010) and in particular very low levels of DNA methylation were detected (Netto et al, 2008). Interestingly, despite the very low DNA methylation level, high levels of the maintenance DNA methyltransferase DNMT1 were found in CIS, indicating that maintenance of DNA methylation has to be on-going (Netto et al, 2008) in the highly proliferating CIS cells (Almstrup et al, 2010).
Upon fertilisation, the maternal and paternal genomes undergo epigenetic reprogramming, mainly characterised by genome-wide loss of DNA methylation. As the only cell type during mammalian development, the PGCs undergo a second round of epigenetic reprogramming where DNA methylation is erased genome-wide. In mice, PGCs at E11.5, which have colonized the gonads, are completely devoid of DNA methylation and remain unmethylated until E13.5 (Hajkova et al, 2002; Seki et al, 2005; Hajkova et al, 2008; Popp et al, 2010; Guibert et al, 2012). DNA methylation of the germ cell genome, including methylation of paternal imprints, is progressively established from E14.5 throughout embryonic development until birth by the de novo DNA methyltransferases (DNMTs) (Kato et al, 2007). Even though less studied, human fetal germ cells seem to show similar re-methylation patterns, where gonocytes, which are un-methylated, at gestational week (GW) 7–17 begin to regain methylation gradually as they mature to pre-spermatogonia (Wermann et al, 2010; Gkountela et al, 2013).
In mammals, the genome-wide DNA demethylation occurs through several interconnected mechanisms, both passive and active. Passive DNA demethylation can occur due to absence or reduction of DNMTs responsible for establishment and maintenance of DNA methylation. Accordingly, Dnmt3a, Dnmt3b and Dnmt1 are significantly downregulated in murine PGCs compared with the neighbouring somatic cells (Seki et al, 2005; Kurimoto et al, 2008). However, the rapid genome-wide DNA demethylation in murine PGCs, within a narrow developmental time window (one day), suggests that additional active demethylation mechanisms exist (Hajkova et al, 2002, 2008).
Two different pathways for active DNA demethylation in PGCs have been proposed. One suggested initiator is the ten eleven translocation (TET) protein family that convert 5mC to hydroxymethylated cytosine (5hmC) (He et al, 2011; Ito et al, 2011). Murine Tet1 and Tet2 are expressed in PGCs, coinciding with the time of rapid 5mC erasure (Hajkova et al, 2010; Hackett et al, 2013; Kagiwada et al, 2013). Another suggested initiating pathway is the direct deamination of 5mC to T by AID/APOBEC1. This introduces a T:G mismatch in the DNA. High expression of Aid and Apobec1 is found in murine PGCs at E10.5–E12.5 (Morgan et al, 2004; Guibert et al, 2012), and Aid-/- PGCs were found to be less demethylated than the wild-type PGCs (Popp et al, 2010). Both of the proposed initiating pathways (TETs and AID/APOBEC1) result in the activation of the base excision repair (BER) machinery, which subsequently to TDG and MBD4 glycosylase activity entails the action of APEX1, PARP1 and XRCC1 (Hegde et al, 2008; Zharkov, 2008). Apex1, Parp1 and Xrcc1 were all found in murine PGCs isolated at E10.5–12.5 where demethylation occurs (Hajkova et al, 2010; Kagiwada et al, 2013).
In contrast to mice, very little is known about DNA demethylation pathways operating in human fetal germ cells. Even less is known about neoplastic germ cells, where this knowledge is of primary importance for understanding the timing of the neoplastic transformation. In the present study, we addressed these gaps in knowledge with focus on CIS. We investigated whether CIS cells, located within the niche of an adult testis, use the same DNA demethylation mechanisms as fetal germ cells, to maintain the low methylation level.
Material and methods
Tissues
The Regional Committee for Medical Research Ethics in Denmark approved the use of human adult and fetal tissue samples from the archives of Copenhagen University Hospital. Adult testicular tissue samples were obtained from testis cancer patients undergoing orchiectomy and from biopsies of the contralateral testis. Tissue samples of normal human fetal testes at GW 11 (where only hypomethylated gonocytes are present), at GW 21 (where a mixture of gonocytes and methylated pre-spermatogonia is present) and at GW 33 (where predominantly pre-spermatogonia are present) were obtained from induced and spontaneous abortions. Gestational age was estimated by the measurement of foot length and sex determined by staining for anti-Müllerian hormone. For the purpose of DNA purification, adult testicular tissue samples were snap-frozen and stored at −80 °C. For immunohistochemistry (IHC) and immunofluorescence (IF) investigations, tissue samples were fixed in formalin or paraformaldehyde at 4 °C overnight immediately after surgery, and subsequently embedded in paraffin.
Testicular-germ-cell-cancer-derived cell lines, NTera2 and TCam-2, were obtained from Dr PW Andrews (UK), and Dr J Shipley (UK), respectively, after permission from Dr S Kitazawa (Japan). Cell lines were cultured according to standard conditions as previously described (Jorgensen et al, 2013). In brief, cells were grown at 37 °C in a 5% CO2 atmosphere, in DMEM (NTera2 cells) or RPMI 1640 (TCam-2 cells) supplemented with 10% fetal calf serum, glutamine (58.5 mg ml−1), penicillin (100 U ml−1) and streptomycin (100 mg ml−1). Cell media and reagents were from Gibco (Naerum, Denmark) and cell culture flasks were from NUNC (Roskilde, Denmark).
RT–PCR
Total RNA was isolated with NucleoSpin RNA II purification kit with DNase I treatment as described by the manufacturer (Macherey-Nagel, Düren, Germany).
The isolated RNA was reverse transcribed with AMV reverse transcriptase (USB, Cleveland, OH, USA) using a dT20 primer and random hexamers and PCR performed using Taq polymerase (GE Healthcare Life Sciences, Freiburg, Germany). The following primers were used:
TET1 (Entrez Gene: 80312) fwd 5′-CACACCAGCTCCACTGAAGA-3′ and rev 5′-CTCCATCACAGGAGCAGACA-3′; TET2 (Entrez Gene: 54790) fwd 5′-CCCACTTACCTGCGTTTCAT-3′ and rev 5′-ACTGTGACCTTTCCCCACTG-3′;
TET3 (Entrez Gene: 200424) fwd 5′-CACCAAGAGTCTGCTGGACA-3′ and rev 5′-CTCCTTCCCCGTGTAGATGA-3′;
PCR conditions were: one cycle of 5 min at 95 °C; 40 cycles of 30 s at 95 °C, 1 min at 62 °C, 1 min at 72 °C and one cycle of 5 min at 72 °C. Products were run on 2% agarose gels and visualised by ethidium bromide staining. Representative bands from each primer combination were excised and sequenced for verification (Eurofins MWG, Ebersberg, Germany).
Laser micro-dissection of CIS
Cryosections of adult testicular tissue samples were prepared, subjected to direct histochemistry and subsequently micro-dissected as previously described (Sonne et al, 2009b; Jorgensen et al, 2011). In brief, tissue samples were embedded in optimum cutting temperature compound (O.C.T. compound, Tissue-Tek, Sakura Finetek Europe, Netherlands), immediately frozen in −78 °C isopentane and stored at −80 °C until use. Sections of 20 μm were cut on a Shandon SME cryotome (Thermo Scientific, Hvidovre, Denmark) and collected on nuclease- and nucleic acid-free membrane slides (Molecular Machines & Industries, Glattbrugg, Switzerland), immediately fixed in 75% RNAse free ethanol and stored in absolute ethanol at −80 °C. To visualise CIS, sections were stained for the activity of alkaline phosphatase with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) solution. Stained CIS cells were micro-dissected from sections within 2 h using the MMI SmartCut System (Olympus). Only tubules with classical appearance of CIS cells along the periphery of tubules were micro-dissected to avoid unspecific stained cells and intratubular tumour cells. For each sample 500–5000 cells were micro-dissected and collected on the adhesive cap of collection tubes (Molecular Machines & Industries, Glattbrugg, Switzerland). Lysis buffer was immediately added to micro-dissected cells and stored at −80 °C.
DNA purification
DNA was purified from tissue samples of embryonal carcinoma (EC; a highly methylated TGCC), tissue samples with CIS situated among normal tubules (CIST), and from micro-dissected CIS cells (MCIS) using the QIAamp DNA Micro Kit (QIAGEN, Copenhagen, Denmark) according to the manufacturer’s protocol. As very little DNA was extracted from samples of micro-dissected CIS, purified DNA was pooled for later application in ELISA. DNA concentration was measured on a NanoDrop ND-1000 Spectrophotometer (Saveen Werner, Limhamn, Sweden).
ELISA
For relative quantification of global content of 5mC and 5hmC, ELISA was performed on DNA purified from EC, CIST and MCIS. ELISA was carried out using the fluorometric assays MethylFlash Methylated DNA Quantification Kit (Colorimetric) and MethylFlash Hydroxymethylated DNA Quantification Kit (Colorimetric) (Epigentek Group Inc., Farmingdale, NY, USA) according to the manufacturer’s protocol. Two hundred nanogram DNA was applied to microtiter plates in three technical replicates and colorimetrical quantification carried out on a microplate spectrophotometer (Tecan Sunrise, Männedorf, Switzerland). The content of 5mC and 5hmC was calculated relative to positive and negative controls included in the kit (containing 50% 5mC and 5hmC, respectively). The assays showed a linear response in the range of 0.1–1% 5mC and 0.01–0.1% 5hmC (at least) and in both cases R2 were >0.9.
Immunohistochemistry
Immunohistochemistry was performed as previously described (Rajpert-De Meyts et al, 2003). A standard indirect peroxidase method was used and development was done primarily with 3-amino-9-ethylcarbazole (yielding a red colour). Sections were counterstained with Mayers Hematoxylin. Primary antibodies were used at indicated dilutions and buffers for microwave treatment as listed in Table 1. For negative controls, serial sections were processed with the primary antibody replaced by the dilution buffer alone. None of the control slides showed any staining (Supplementary Figure 1). Positive controls were samples of intestine (for APOBEC1), prostate (for TET1), EC (for DNMTs) and normal testis (the remaining antibodies). In all cases controls stained positive (data not shown).
Sections were scanned on NanoZoomer (Hamamatsu Photonics, Herrsching am Ammersee, Germany) and NDP.view software (Hamamatsu Photonics) was used for analysis. Tubules containing CIS cells were identified in serial sections by staining with classical CIS markers D2-40/M2A/PDPN (Marks et al, 1999; Sonne et al, 2006b) or PLAP (Giwercman et al, 1991). For each tissue sample, the intensity of the staining of CIS cells, was evaluated and scored as negative (neg), faint staining (+), moderate staining (++) and strongly positive staining (+++). The general staining pattern observed across the investigated tissue samples was subsequently summarised, by collectively describing all the staining intensities observed in the included samples. If staining was observed only in a few (1–2) of the included tissue samples, this was noted (few). Tissue samples were excluded from the summary if the following was observed: abnormal morphology (probably due to bad tissue preservation), lack of staining in a positive control and increased background staining.
Immunofluorescence double staining
Immunofluorescence double staining was performed on adult and fetal testicular tissue samples. The protocol described above for IHC was used for IF with only minor adjustments. Primary antibodies were applied overnight at 4 °C and left for 1 h at room temperature (RT) the following day. Fetal gonocytes were detected by OCT3/4, pre-spermatogonia were detected by a spermatogonial marker MAGE-A4 (Aubry et al, 2001; Gaskell et al, 2004) and CIS cells identified by the specific markers D2-40 or PLAP as described above. Primary antibodies were used at indicated dilutions and buffers as listed in Table 1. For negative controls, dilution buffer without the primary antibody was applied to serial sections. No staining was observed in negative control samples (Figure 1 and Supplementary Figure 1). The following secondary antibodies were applied in pairs according to the respective primary antibodies: Alexa Fluor 488 goat anti-rabbit, Alexa Fluor 488 donkey anti-goat, Alexa Fluor 568 donkey anti-mouse (Life Technologies, Naerum, Denmark), all diluted 1 : 600, and incubation took place for 45 min at RT. Sections were counterstained with DAPI (2 μg ml−1 in TBS), mounted with anti-fade mounting solution (Prolong Gold anti-fade reagent, P36930, Life Technologies) and left to dry for 24 h. In between incubation steps, slides were washed in TBS. Finally, cover glasses were sealed with nail polish. Fluorescence microscopy analysis was carried out on Olympus BX61 (Olympus Ltd., Southend-on-Sea, UK) coupled to the fluorescent light source X-Cite series 120 PC Q (Lumen Dynamics Group Inc., Mississauga, ON, Canada). Image processing, acquisition and photography were performed with the software cellSens Dimensions version 1.6 (Olympus Ltd.).
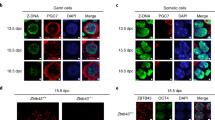
Assessment of 5mC and 5hmC levels in CIS cells and IF detection of TET proteins. (A, C) Quantitative measurements of respectively the 5mC and 5hmC levels in DNA isolated from micro-dissected CIS cells (MCIS and MCIS/NS), whole tissue section of testicular tissue containing CIS (CIST) and tissue containing embryonal carcinoma (EC). The content of 5mC and 5hmC was calculated relative to positive and negative controls included in the kit (containing 50% 5mC and 5hmC, respectively). Bars represent mean±std. (B, D) Immunohistochemical staining for, respectively, 5mC and 5hmC in testicular tissue with CIS cells (arrowheads). Bars represent 25 microns. (E) Immunofluorescence detection of TET1 (upper row of images) and TET2 (the bottom row) is shown in green. CIS cells (arrowheads) are marked with D2-40 (red) and DAPI (blue) stains in nuclei. Bars represent 10 μm. Negative control samples are shown in Supplementary Figure 1. TET1 and TET2 expression in normal testis are shown in Supplementary Figure 2B.
Western blotting
Western blotting (WB) was performed in order to validate the specificity of antibodies used in this study (Table 1). Protein was extracted from frozen testicular tissue samples containing CIS cells in all tubules (CIS+NT), normal adult testis (NT), seminoma (s.e.m.), EC and from the cell lines TCam2 and NT2. Samples were prepared and WB performed as previously described (Sonne et al, 2006a), except that HRP-conjugated secondary antibodies were used and protein signal detected by chemiluminescent detection with Pierce ECL detection (No. 34095, Thermo Scientific). Ten microgram of protein was loaded in each lane and primary antibodies were diluted 1 : 200 in 1% milk powder with TBS Tween (0.1%), except TET1 which was diluted 1 : 100. β-actin was used as loading control and the primary antibody diluted 1 : 200 (β-actin, Santa Cruz Sc-47778, Santa Cruz, CA, USA). Secondary antibodies were HRP-conjugated rabbit anti-mouse (P0260, Dako, Glostrup, Denmark), HRP-conjugated rabbit anti-goat (P0160, Dako) and HRP-conjugated swine anti-rabbit (P0217, Dako), all diluted at 1 : 1000. For all antibodies investigated protein bands of expected size were detected (see Supplementary Figure 2).
Results
Are CIS cells hydroxymethylated by TET proteins?
The TET family proteins are responsible for the conversion of 5mC to 5hmC. TET proteins and 5hmC have been identified in PGCs and may have a pivotal role in demethylation of their genome during development. Owing to the close resemblance of CIS to fetal germ cells (Almstrup et al, 2004; Sonne et al, 2009a; Almstrup et al, 2010), we asked whether TET proteins and 5hmC might be present in CIS cells. We therefore performed IHC of 5hmC and 5mC (Figure 1B and D) and found no staining in the nuclei of CIS cells, whereas intense staining was present in the nuclei of the adjacent Sertoli cells. To address the question further, we isolated CIS cells from testicular tissue sections by laser micro-dissection and purified the DNA. By ELISA, we assessed the amount of 5hmC and 5mC in DNA obtained from two different micro-dissected samples (one with CIS next to a non-seminoma: MCIS/NS; and one with CIS without tumour: MCIS) as well as from a whole tissue sample containing testicular tissue with CIS (CIST) and a sample with EC (which is highly methylated). Both of the micro-dissected samples showed low levels of 5hmC and 5mC (Figure 1A and C), in accordance with our findings by IHC. We therefore conclude that CIS cells retain low levels of both 5mC and 5hmC. The EC sample and the sample with CIS from a whole tissue section showed substantially higher levels of both 5mC and 5hmC.
RT–PCR of RNA from biopsies containing CIS and different TGCTs, as well as TGCT-derived cell lines, showed that TET1 and TET2 were found in nearly all samples, whereas TET3 was absent from tissue containing CIS cells (Supplementary Figure 3A). We hence focused on TET1 and TET2, which are also described to be the TET proteins mostly engaged in 5mC hydroxylation (Koh et al, 2011). As shown in Figure 1E, we neither detected TET1 nor TET2 in the nucleus of CIS cells by IF. A slight staining in the cytoplasm of CIS cells could however not be excluded. TET1 and TET2 were, however, detected in spermatogonia, spermatocytes and Leydig cells (Supplementary Figure 3B), which explains the presence of detectable messenger RNA by RT–PCR in tissue blocks containing various kinds of testicular cells. Interrogation of earlier microarray data on micro-dissected cell populations (Sonne et al, 2009a), also indicated a low expression of all TET proteins (Supplementary Figure 3C). We conclude that the TET proteins are not expressed in the nucleus of CIS cells and that only very low levels of 5hmC can be detected in the genome of CIS cells.
Are CIS cells engaged in active DNA demethylation?
We next asked whether proteins that have been described to be engaged in active DNA demethylation in PGCs also were present in CIS cells. By IHC, we investigated APOBEC1, MBD4, APEX1 and PARP1 and found all to be present in the nuclei of CIS cells (Table 2). XRCC1 was not found in CIS cells (Supplementary Figure 4). As shown in Figure 2, APOBEC1, MBD4, APEX1 and PARP1 all co-localised to CIS cells marked by either PLAP or D2-40. APOBEC1 localised mainly to the cytoplasm, but less intense staining was also found in the nucleus. MBD4, APEX1 and PARP1 were all localised to the nuclei of CIS cells. We conclude that CIS cells seem to express proteins that facilitate active DNA demethylation by AID/APOBEC1 and BER.
Immunofluorescence detection of proteins involved in active DNA demethylation in adult testis samples with CIS. On the left-hand side differential interference contrast (DIC) images display morphology of each section and are merged with DAPI staining (blue) to reveal nuclei. CIS cells (arrowheads) are marked by the classical CIS markers PLAP or D2-40 (red). The CIS cells show expression of the markers of active demethylation APOBEC1, MBD4, APEX1 and PARP1 (green). Bars represent 10 μm.
Do human fetal germ cells express the same DNA demethylation markers as CIS cells?
As a high degree of similarity between gonocytes and CIS cells has been described (recently reviewed in Kristensen et al, 2013), we next asked whether human fetal germ cells express the same DNA demethylation proteins as CIS cells. By IF, we investigated fetal germ cells at GW 11, 21 and 33 for the co-localisation of APOBEC1, MBD4, APEX1 and PARP1. Fetal germ cells were identified by OCT3/4 at GW 11 and by MAGE-A4 at GW 21 and 33, both specifically mark fetal germ cells at the developmental stages corresponding to gonocytes and pre-spermatogonia, respectively (Gaskell et al, 2004). At GW 21, we found MBD4 and PARP1 to be highly expressed in the nucleus of fetal germ cells (Figure 3), as high intensities of fluorescent signals were observed. APOBEC1 and APEX1 localised to the cytoplasm of fetal germ cells, and were expressed at a lower level compared with MBD4 and PARP1, as lower intensities of fluorescent signals were observed. APOBEC1, MBD4, APEX1 and PARP1 also localised to MAGE-A4 negative cells within the tubules, which could be fetal germ cells at an earlier developmental stage, most likely gonocytes. In addition, some variation was observed for MBD4 expression, as germ cells from the same section could be found both positive and negative. The same observations were found for fetal testis samples at GW 11 and GW 33 (Supplementary Figure 5), except that APOBEC1 also localised to the nuclear membrane of fetal germ cells at GW 33, in addition to the previously described cytoplasmic localisation. We conclude that human fetal germ cells express many of the proteins that facilitate active DNA demethylation by AID/APOBEC1 and BER, but at an apparent lower level than CIS cells and with some variation.
Immunofluorescence detection of proteins involved in active DNA demethylation in a human fetal testis sample at 21 weeks of gestation (GW). A differential interference contrast (DIC) image displaying morphology of each section merged with DAPI (blue) staining to reveal nuclei, is shown to the left. Human fetal germ cells (arrowheads) marked by MAGE-A4 (red) show expression of most of the markers of active demethylation APOBEC1, MBD4, APEX1 and PARP1 (green). Bars represent 10 μm.
Are CIS cells also engaged in passive DNA demethylation?
Lastly we asked whether the genome of CIS cells might also be demethylated passively as a consequence of absent or down-regulated DNMTs. By IHC, we investigated the expression of DNMT1, DNMT3A, DNMT3B and DNMT3L. We found DNMT1, DNMT3B and DNMT3L localised in the nucleus of CIS cells, whereas DNMT3A was absent (Table 2). By IF, we showed that DNMT1 co-localised to the nuclei of CIS cells. DNMT3B and its co-enzyme DNMT3L were also found to co-localise to CIS cells, but were expressed at a lower level and, very interestingly, in small foci within the nuclei (Figure 4 and Supplementary Figure 6). Some variation was observed among CIS cells from the same section with regard to expression of DNMT3B and L (Figure 4). DNMT3A was not observed in CIS, but was present in the nuclei of Sertoli cells. We conclude that the maintenance methyl-transferase DNMT1 is expressed in CIS cells and that de novo methyl-transferases DNMT3B and 3L also could be detected in the nucleus of CIS cells; however, the level seemed lower and some variation was observed.
Adult testis samples with CIS displaying IF detection of DNMT proteins (green) involved in generation of 5mC. On the left-hand side differential interference contrast (DIC) images display morphology of each section and are merged with DAPI staining (blue) to reveal nuclei. CIS cells are marked with D2-40 (arrowheads). CIS cells showed a substantial expression of DNMT1. DNMT3B and L show a faint expression in small foci in the nucleus of CIS cells (high-power magnification is shown in Supplementary Figure 6). DNMT3A showed no expression in CIS cells. Bars represent 10 μm.
Discussion
We have previously shown that CIS cells retain an open chromatin and gonocyte-like epigenetic modifications (Almstrup et al, 2010). In addition to the permissive chromatin observed in CIS cells, one of the most noteworthy epigenetic features of CIS cells is the complete absence or extremely low level of 5mC (Smiraglia et al, 2002; Netto et al, 2008). Human fetal germ cells are also hypomethylated during the embryonic period, but gradually become methylated (5mC positive immunostaining) and by the time of birth most germ cells are methylated (Wermann et al, 2010). A hypomethylated genome promotes proliferation and may cause genomic instability thereby contributing to tumorigenesis (Eden et al, 2003). DNA hypomethylation may also facilitate aberrant expression of pluripotency genes, like POU5F1, NANOG and TFAP2C, which indeed are highly expressed in CIS cells (Looijenga et al, 2003; Hoei-Hansen et al, 2004; Rajpert-De Meyts et al, 2004; Hoei-Hansen et al, 2005). Aberrant expression of other epigenetic modifiers in CIS cells, like BLIMP/PRMT5 (Eckert et al, 2008), might also add to abnormal epigenetic reprogramming and thereby genomic instability.
In this study, we show that CIS cells express APOBEC1, MBD4, APEX1 and PARP1, proteins that are involved in active removal of 5mC. A possible pitfall is however that many of these proteins have dual or multiple roles. MBD4, APEX1 and PARP1 are part of the BER complex, which functions also in repair of DNA besides being engaged in active removal of 5mC. The presence of BER components in CIS cells does not per se prove that they participate in retaining the genome in a hypomethylated state. However, we also showed that the known alternative demethylation pathway via TET proteins and the generation of 5hmC was absent in CIS cells, as we observed very low levels of 5hmC and no expression of TET1 and TET2 in the nucleus of CIS cells. This stands in contrast to studies in mice, where TETs are suggested to be the main proteins involved in the demethylation of the genome in PGCs, based on observations of an initial increase (E10.5–E11.5) and a subsequent decrease (E13.5) in 5hmC levels, coinciding with decreasing 5mC levels (Hackett et al, 2013). However, knock out of Tet1 in mouse did not impair fertility and only resulted slightly increased methylation levels (Yamaguchi et al, 2012). Hence, even though it seems likely that 5hmC and TET proteins have an important role during fetal germ cell development, Tet1 is dispensable in the development of normal murine germ cells.
Our results tend to suggest an active demethylation pathway initiated by the deaminase APOBEC1 and not TETs, as CIS cells expressed APOBEC1 and the downstream BER components MBD4, APEX1 and PARP1, where the latter indeed have been demonstrated to be involved in active DNA demethylation in murine PGCs (Morgan et al, 2004; Hajkova et al, 2010; Popp et al, 2010; Hashimoto et al, 2012; Nabel et al, 2012). However, contradictory results regarding the level of expression of APOBEC1 in murine PGCs have been reported (Hajkova et al, 2010; Kagiwada et al, 2013).
Inhibition of DNMTs is yet another alternative pathway to hypomethylate a genome. Replication-coupled passive DNA demethylation has been suggested to be sufficient for demethylation of the genome in murine PGCs, as they were found to divide more rapidly than previously thought and to have a low expression of DNMTs, leading to dilution of 5mC over time (Kagiwada et al, 2013). Even though CIS cells are highly proliferative and express BLIMP1 (Eckert et al, 2008; Almstrup et al, 2010), we did find a substantial expression of DNMT1, which is in line with a previous report (Netto et al, 2008) and low expression of DNMT3B and L in CIS cells. Hence, both maintenance and de novo methyltransferases seem to be present in CIS cells, but it is yet unknown whether they are post-translationally inhibited or whether the low level of de novo DNMTs is sufficient to drive re-methylation of the CIS genome.
In any case, multiple mechanisms are likely to operate in concert to demethylate the genome of CIS cells and fetal gonocytes. As in CIS cells, we found APOBEC1, MBD4, APEX1 and PARP1 to be expressed in human male fetal germ cells at GW11, GW21 and GW33, when a transition from hypomethylated gonocytes to methylated pre-spermatogonia is in progress (Wermann et al, 2010; Gkountela et al, 2013). The localisation of APOBEC1 to the cytoplasm of fetal germ cells at GW11 and GW21, could indicate that it may not be involved in active DNA demethylation. The appearance of APOBEC1 in the nuclear membrane of fetal germ cells at GW33, could indicate a function of APOBEC1 in active DNA demethylation in the nucleus at this later time point. The presence of APOBEC1 in human gonocytes stands in contrast to the absence of APOBEC1 observed in murine PGCs (Hajkova et al 2010). However, epigenetic cues in human and murine fetal germ cells may differ (Almstrup et al, 2010; Gkountela et al, 2013). In particular, a more prolonged process of demethylation is evident in human fetal germ cells compared with the rapid demethylation in murine PGCs (Wermann et al, 2010; Guibert et al, 2012).
In this study, we found that PARP1 was expressed in the nuclei of CIS cells. In addition to its role in BER, PARP1 participates in regulating the activity of a wide range of proteins involved in epigenetic modulation (Quenet et al, 2009; Gibson and Kraus, 2012). When bound to its target, PARP1 synthesizes a negatively charged polymer of poly-(ADP-ribose) units, which can be added to Glu, Asp or Lys residues of target proteins. Thereby, PARP1 performs posttranscriptional modification of its target proteins – a process also called PARylation. PARP1 can thereby affect the chromatin structure and has been shown to impair chromatin remodelling, histone demethylation and nucleosome–nucleosome interaction facilitating decondensation of chromatin (Gibson and Kraus, 2012). It is hence possible, that PARP1 itself can contribute to an unstable chromatin structure in CIS and fetal germ cells, in addition to its role in active demethylation. The high expression of PARP1 in CIS is supported by our earlier results on the CIS chromatin, which we found to be in a highly open and permissive conformation (Almstrup et al, 2010).
Epigenetic modifications occurring during embryonic development are well recognised as being susceptible to alterations by exposure to environmental factors. Indeed, animal studies show that certain exposures (e.g. to endocrine disrupting chemicals) during fetal development cause changes in epigenetic patterns with a phenotypic outcome, including alterations in the development of male germ cells (Feil and Fraga, 2011). Studies in humans have shown that e.g. prenatal starvation can be traced in the adult epigenome of peripheral blood mononuclear cells affecting, for example, the insulin promoter (Heijmans et al, 2008; Tobi et al, 2009). The important role of epigenetics during development and experimental evidence that environmental factors can influence the epigenetic programming indicate a role for epigenetics in the pathogenesis of TGCC. It is speculative at this point but likely that in utero environmental exposure can affect the reprogramming of the epigenome in developing fetal germ cells and thereby contribute to their neoplastic transformation into CIS. The present study substantiates this viewpoint by indicating developmental failure to end active DNA demethylation processes in CIS cells. Cues to inhibit demethylation processes in fetal germ cells could indeed be promoted by maturation of surrounding Sertoli cells and hence we speculate that the lack of this function may be a consequence of testicular dysgenesis or an under-virilised gonad as suggested earlier by our group (Skakkebaek et al, 2001).
In conclusion, the common precursor of TGCC – CIS – was shown to be devoid of TET proteins and with low levels of both 5mC and 5hmC. In contrast, the hypomethylated CIS cells expressed a range of proteins that have been implicated in active demethylation of fetal germ cells. We therefore suggest that the CIS genome is subjected to active DNA demethylation. The hypomethylated state is likely associated with a continuous expression of pluripotency genes and may facilitate increased proliferation of CIS cells exposed to a high hormonal activity of the post-pubertal testis, which are features that undoubtedly contribute to the invasive progression.
Change history
04 February 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Albrechtsen R, Nielsen M, Skakkebaek N, Wewer U (1982) Carcinoma in situ of the testis. Some ultrastructural characteristics of germ cells. Acta Pathol Microbiol Immunol Scand A 90 (4): 301–303.
Almstrup K, Hoei-Hansen C, Wirkner U, Blake J, Schwager C, Ansorge W, Nielsen J, Skakkebaek N, Rajpert-De Meyts E, Leffers H (2004) Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res 64 (14): 4736–4743.
Almstrup K, Leffers H, Lothe R, Skakkebaek N, Sonne S, Nielsen J, Rajpert-De Meyts E, Skotheim R (2007) Improved gene expression signature of testicular carcinoma in situ. Int J Androl 30 (4): 292–302, discussion 303.
Almstrup K, Nielsen JE, Mlynarska O, Jansen MT, Jorgensen A, Skakkebaek NE, Rajpert-De Meyts E (2010) Carcinoma in situ testis displays permissive chromatin modifications similar to immature foetal germ cells. Br J Cancer 103 (8): 1269–1276.
Aubry F, Satie AP, Rioux-Leclercq N, Rajpert-De Meyts E, Spagnoli GC, Chomez P, De Backer O, Jegou B, Samson M (2001) MAGE-A4, a germ cell specific marker, is expressed differentially in testicular tumors. Cancer 92 (11): 2778–2785.
Berthelsen JG, Skakkebaek NE, Mogensen P, Sørensen BL (1979) Incidence of carcinoma in situ of germ cells in contralateral testis of men with testicular tumours. Br Med J 2 (6186): 363–364.
Biermann K, Heukamp L, Steger K, Zhou H, Franke F, Guetgemann I, Sonnack V, Brehm R, Berg J, Bastian P, Müller S, Wang-Eckert L, Schorle H, Büttner R (2007) Gene expression profiling identifies new biological markers of neoplastic germ cells. Anticancer Res 27 (5A): 3091–3100.
Chia V, Quraishi S, Devesa S, Purdue M, Cook M, McGlynn K (2010) International trends in the incidence of testicular cancer, 1973-2002. Cancer epidemiol Biomarkers Prevent 19 (5): 1151–1159.
Chung CC, Kanetsky PA, Wang Z, Hildebrandt MA, Koster R, Skotheim RI, Kratz CP, Turnbull C, Cortessis VK, Bakken AC, Bishop DT, Cook MB, Erickson RL, Fossa SD, Jacobs KB, Korde LA, Kraggerud SM, Lothe RA, Loud JT, Rahman N, Skinner EC, Thomas DC, Wu X, Yeager M, Schumacher FR, Greene MH, Schwartz SM, McGlynn KA, Chanock SJ, Nathanson KL (2013) Meta-analysis identifies four new loci associated with testicular germ cell tumor. Nat Genet 45 (6): 680–685.
Eckert D, Biermann K, Nettersheim D, Gillis A, Steger K, Jäck H, Müller A, Looijenga L, Schorle H (2008) Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev Biol 8: 106 doi: 10.1186/1471-213X-8-106.
Eden A, Gaudet F, Waghmare A, Jaenisch R (2003) Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300 (5618): 455.
Feil R, Fraga MF (2011) Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 13 (2): 97–109.
Gaskell TL, Esnal A, Robinson LL, Anderson RA, Saunders PT (2004) Immunohistochemical profiling of germ cells within the human fetal testis: identification of three subpopulations. Biol Reprod 71 (6): 2012–2021.
Gibson BA, Kraus WL (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13 (7): 411–424.
Giwercman A, Cantell L, Marks A (1991) Placental-like alkaline phosphatase as a marker of carcinoma-in-situ of the testis. Comparison with monoclonal antibodies M2A and 43-9F. APMIS 99 (7): 586–594.
Gkountela S, Li Z, Vincent JJ, Zhang KX, Chen A, Pellegrini M, Clark AT (2013) The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol 15 (1): 113–122.
Guibert S, Forne T, Weber M (2012) Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 22 (4): 633–641.
Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA (2013) Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339 (6118): 448–452.
Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange U, Cesari F, Lee C, Almouzni G, Schneider R, Surani M (2008) Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452 (7189): 877–881.
Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani M (2002) Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117 (1-2): 15–23.
Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA (2010) Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329 (5987): 78–82.
Hashimoto H, Zhang X, Cheng X (2012) Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation. Nucleic Acids Res 40 (17): 8276–8284.
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333 (6047): 1303–1307.
Hegde ML, Hazra TK, Mitra S (2008) Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res 18 (1): 27–47.
Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH (2008) Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA 105 (44): 17046–17049.
Hemminki K, Li X (2002) Cancer risks in second-generation immigrants to Sweden. Int J Cancer 99 (2): 229–237.
Hoei-Hansen C, Almstrup K, Nielsen J, Brask Sonne S, Graem N, Skakkebaek N, Leffers H, Rajpert-De Meyts E (2005) Stem cell pluripotency factor NANOG is expressed in human fetal gonocytes, testicular carcinoma in situ and germ cell tumours. Histopathology 47 (1): 48–56.
Hoei-Hansen C, Nielsen J, Almstrup K, Sonne S, Graem N, Skakkebaek N, Leffers H, Rajpert-De Meyts E (2004) Transcription factor AP-2gamma is a developmentally regulated marker of testicular carcinoma in situ and germ cell tumors. Clin Cancer Res 10 (24): 8521–8530.
Huyghe E, Matsuda T, Thonneau P (2003) Increasing incidence of testicular cancer worldwide: a review. J Urol 170 (1): 5–11.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333 (6047): 1300–1303.
Jorgensen A, Blomberg Jensen M, Nielsen JE, Juul A, Rajpert-De Meyts E (2013) Influence of vitamin D on cisplatin sensitivity in testicular germ cell cancer-derived cell lines and in a NTera2 xenograft model. J Steroid Biochem Mol Biol 136: 238–246.
Jorgensen A, Dalgaard MD, Sonne SB (2011) Microdissection of gonadal tissues for gene expression analyses. Methods Mol Biol 755: 307–313.
Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M (2013) Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 32 (3): 340–353.
Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H (2007) Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16 (19): 2272–2280.
Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A (2011) Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8 (2): 200–213.
Kristensen DG, Skakkebaek NE, Rajpert-De Meyts E, Almstrup K (2013) Epigenetic features of testicular germ cell tumours in relation to epigenetic characteristics of foetal germ cells. Int J Dev Biol 57 (2-4): 309–317.
Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M (2008) Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev 22 (12): 1617–1635.
Looijenga L, Stoop H, de Leeuw H, de Gouveia Brazao C, Gillis A, van Roozendaal K, van Zoelen E, Weber R, Wolffenbuttel K, van Dekken H, Honecker F, Bokemeyer C, Perlman E, Schneider D, Kononen J, Sauter G, Oosterhuis J (2003) POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res 63 (9): 2244–2250.
Marks A, Sutherland DR, Bailey D, Iglesias J, Law J, Lei M, Yeger H, Banerjee D, Baumal R (1999) Characterization and distribution of an oncofetal antigen (M2A antigen) expressed on testicular germ cell tumours. Br J Cancer 80 (3-4): 569–578.
Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK (2004) Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem 279 (50): 52353–52360.
Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM (2012) AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol 8 (9): 751–758.
Netto G, Nakai Y, Nakayama M, Jadallah S, Toubaji A, Nonomura N, Albadine R, Hicks J, Epstein J, Yegnasubramanian S, Nelson W, De Marzo A (2008) Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod Pathol 21 (11): 1337–1344.
Nielsen H, Nielsen M, Skakkebaek N (1974) The fine structure of possible carcinoma-in-situ in the seminiferous tubules in the testis of four infertile men. Acta Pathol Microbiol Scand A 82 (2): 235–248.
Novotny GW, Belling KC, Bramsen JB, Nielsen JE, Bork-Jensen J, Almstrup K, Sonne SB, Kjems J, Rajpert-De Meyts E, Leffers H (2012) MicroRNA expression profiling of carcinoma in situ cells of the testis. Endocr Relat Cancer 19 (3): 365–379.
Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463 (7284): 1101–1105.
Quenet D, El Ramy R, Schreiber V, Dantzer F (2009) The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol 41 (1): 60–65.
Rajpert-De Meyts E (2006) Developmental model for the pathogenesis of testicular carcinoma in situ: genetic and environmental aspects. Hum Reprod 12 (3): 303–323.
Rajpert-De Meyts E, Bartkova J, Samson M, Hoei-Hansen C, Frydelund-Larsen L, Bartek J, Skakkebaek N (2003) The emerging phenotype of the testicular carcinoma in situ germ cell. APMIS 111 (1): 267–278, discussion 278-9.
Rajpert-De Meyts E, Hanstein R, Jorgensen N, Graem N, Vogt PH, Skakkebaek NE (2004) Developmental expression of POU5F1 (OCT-3/4) in normal and dysgenetic human gonads. Hum Reprod 19 (6): 1338–1344.
Ruark E, Seal S, McDonald H, Zhang F, Elliot A, Lau K, Perdeaux E, Rapley E, Eeles R, Peto J, Kote-Jarai Z, Muir K, Nsengimana J, Shipley J, Bishop DT, Stratton MR, Easton DF, Huddart RA, Rahman N, Turnbull C (2013) Identification of nine new susceptibility loci for testicular cancer, including variants near DAZL and PRDM14. Nat Genet 45 (6): 686–689.
Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y (2005) Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278 (2): 440–458.
Skakkebaek N (1972) Possible carcinoma-in-situ of the testis. Lancet 2 (7776): 516–517.
Skakkebaek N, Berthelsen J, Giwercman A, Müller J (1987) Carcinoma-in-situ of the testis: possible origin from gonocytes and precursor of all types of germ cell tumours except spermatocytoma. Int J Androl 10 (1): 19–28.
Skakkebaek NE, Rajpert-De Meyts E, Main KM (2001) Testicular dysgenesis syndrome: an increasingly common developmental disorder with environmental aspects. Hum Reprod 16 (5): 972–978.
Skotheim R, Lind G, Monni O, Nesland J, Abeler V, Fosså S, Duale N, Brunborg G, Kallioniemi O, Andrews P, Lothe R (2005) Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res 65 (13): 5588–5598.
Smiraglia D, Szymanska J, Kraggerud S, Lothe R, Peltomäki P, Plass C (2002) Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene 21 (24): 3909–3916.
Sonne S, Almstrup K, Dalgaard M, Juncker A, Edsgard D, Ruban L, Harrison N, Schwager C, Abdollahi A, Huber P, Brunak S, Gjerdrum L, Moore H, Andrews P, Skakkebaek N, Meyts E, Leffers H (2009a) Analysis of gene expression profiles of microdissected cell populations indicates that testicular carcinoma in situ is an arrested gonocyte. Cancer Res 69 (12): 5241–5250.
Sonne S, Hoei-Hansen C, Nielsen J, Herlihy A, Andersson A, Almstrup K, Daugaard G, Skakkebaek N, Leffers H, Rajpert-De Meyts E (2006a) CDH1 (E-cadherin) in testicular germ cell neoplasia: suppressed translation of mRNA in pre-invasive carcinoma in situ but increased protein levels in advanced tumours. APMIS 114 (7-8): 549–558.
Sonne SB, Dalgaard MD, Nielsen JE, Hoei-Hansen CE, Rajpert-De Meyts E, Gjerdrum LM, Leffers H (2009b) Optimizing staining protocols for laser microdissection of specific cell types from the testis including carcinoma in situ. PLoS One 4 (5): e5536.
Sonne SB, Herlihy AS, Hoei-Hansen CE, Nielsen JE, Almstrup K, Skakkebaek NE, Marks A, Leffers H, Rajpert-De Meyts E (2006b) Identity of M2A (D2-40) antigen and gp36 (Aggrus, T1A-2, podoplanin) in human developing testis, testicular carcinoma in situ and germ-cell tumours. Virchows Arch 449 (2): 200–206.
Sperger J, Chen X, Draper J, Antosiewicz J, Chon C, Jones S, Brooks J, Andrews P, Brown P, Thomson J (2003) Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci USA 100 (23): 13350–13355.
Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT (2009) DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet 18 (21): 4046–4053.
Turnbull C, Rahman N (2011) Genome-wide association studies provide new insights into the genetic basis of testicular germ-cell tumour. Int J Androl 34 (4 Pt 2): e86–e96, discussion e96-7.
Wermann H, Stoop H, Gillis AJ, Honecker F, van Gurp RJ, Ammerpohl O, Richter J, Oosterhuis JW, Bokemeyer C, Looijenga LH (2010) Global DNA methylation in fetal human germ cells and germ cell tumours: association with differentiation and cisplatin resistance. J Pathol 221 (4): 433–442.
Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y (2012) Tet1 controls meiosis by regulating meiotic gene expression. Nature 492 (7429): 443–447.
Zharkov DO (2008) Base excision DNA repair. CMLS 65 (10): 1544–1565.
Acknowledgements
The work herein was supported by the Research Fund of Rigshospitalet (to JEN, no. 9615.06.1.15), the Lundbeck Foundation (to ERM, no. R48-A4746) and The Danish Cancer Society (to ERM, no. R40-A2127-B1276). We are very grateful for the skilled technical work by Ana Ricci Nielsen, Brian Vendelbo Hansen and Betina F Nielsen. We thank Dr G Spagnoli for a generous gift of MAGE-A4 antibody, Dr K Okamoto for the generous gift of DNMT3L antibody, Dr PW Andrews for NTera2 cells, and Drs S Kitazawa and J Shipley for TCam-2 cells.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Kristensen, D., Nielsen, J., Jørgensen, A. et al. Evidence that active demethylation mechanisms maintain the genome of carcinoma in situ cells hypomethylated in the adult testis. Br J Cancer 110, 668–678 (2014). https://doi.org/10.1038/bjc.2013.727
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.727
Keywords
This article is cited by
-
Epigenetic drugs and their molecular targets in testicular germ cell tumours
Nature Reviews Urology (2019)
-
Testicular germ cell tumor: a comprehensive review
Cellular and Molecular Life Sciences (2019)
-
DNA hypomethylation and aberrant expression of the human endogenous retrovirus ERVWE1/syncytin-1 in seminomas
Retrovirology (2017)
-
Clinical observations on chemotherapy curable malignancies: unique genetic events, frozen development and enduring apoptotic potential
BMC Cancer (2015)
-
Profiling of the small RNA populations in human testicular germ cell tumors shows global loss of piRNAs
Molecular Cancer (2015)