Abstract
Background:
Spinophilin, a multifunctional intracellular scaffold protein, is reduced in certain types of cancer and is regarded as a novel putative tumour suppressor protein. However, the role of spinophilin in hepatocellular carcinoma (HCC) has never been explored before.
Methods:
In this study, we determined for the first time the expression pattern of spinophilin in human HCC by immunohistochemistry and quantitative reverse transcriptase–PCR analysis. In addition, we performed immunohistochemical analysis of p53, p14ARF and the proliferation marker Ki-67. Kaplan–Meier curves and multivariate Cox proportional models were used to study the impact on clinical outcome. Small interfering RNA (siRNA) was used to silence spinophilin and to explore the effects of reduced spinophilin expression on cellular growth.
Results:
In our study, complete loss of spinophilin immunoreactivity was found in 44 of 104 HCCs (42.3%) and reduced levels were found in an additional 37 (35.6%) cases. After adjusting for other prognostic factors, multivariate Cox regression analysis identified low expression of spinophilin as an independent prognostic factor with respect to disease-free (hazard ratio (HR)=1.8; 95% confidence interval (CI)=1.04–3.40; P=0.043) and cancer-specific survival (HR=2.0; CI=1.1–3.8; P=0.025). Reduced spinophilin expression significantly correlated with higher Ki-67 index in HCC (P=0.014). Reducing spinophilin levels by siRNA induced a higher cellular growth rate and increased cyclin D2 expression in tumour cells (P<0.05).
Conclusion:
This is the first study of the expression pattern and distribution of spinophilin in HCC. According to our data, the loss of spinophilin is associated with higher proliferation and might be useful as a prognostic marker in patients with HCC.
Similar content being viewed by others
Main
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortality worldwide (Siegel et al, 2012). Owing to an increased prevalence in causal risk factors, such as obesity and viral liver infections, the incidence of HCC is increasing in developed countries (Shariff et al, 2009). Despite the introduction of novel agents, including the multikinase inhibitor sorafenib for metastatic HCC, the disseminated disease stage is incurable. For locally confined HCC, resection is the gold standard in the curative treatment of this disease (Frau et al, 2010). However, many patients fail a successful complete resection or develop a recurrence in other regions of the organ (Lai et al, 2012). Currently, because of the heterogeneity of the underlying molecular pathophysiological mechanisms in the tumour as well as in the non-neoplastic transformed adjacent liver (Guichard et al, 2012), the widely used TNM staging classification system is limited in predicting the course of the disease, the recurrence rate and the survival of patients with HCC (Pons et al, 2005). Therefore, novel prognostic factors are of paramount importance to classify adequately HCC patients with regard to different treatment modalities, to predict individual outcomes, to report pathologic specimens, to conduct postoperative surveillance and to qualify for inclusion into multinational epidemiologic, exploratory and interventional clinical trials (Minguez and Lachenmayer, 2011). Recently, many studies have attempted to evaluate clinicopathological features or discover novel biomarkers that lead to a better prediction of the clinical course of HCC (Lauwers et al, 2002; Aigelsreiter et al, 2009, 2012). In general, the detailed characterisation of the biological properties of a particular pathophysiological factor could provide a logical rationale for testing its potential prognostic significance as a biomarker in HCC. In this context, spinophilin (Spn; also known as Neurabin 2), a multifunctional protein whose gene is located in the 17q21.33 chromosomal region, is commonly downregulated or lost in different human cancers, including renal cell carcinoma, adenocarcinoma of the lung and tumours of the central nervous system (Carnero, 2012). Furthermore, lower levels of Spn mRNA have been correlated with higher grades of ovarian carcinoma and the progression of chronic myelogenous leukaemia to a more aggressive phenotype (Carnero, 2012). As a consequence, Spn has been proposed to be a novel tumour suppressor in human cancers (Carnero, 2012). Notably, its chromosomal location contains a relatively high density of well-known tumour suppressor genes and is commonly impaired by chromosomal alterations in HCCs (Furge et al, 2005). To date, no study has been published that has explored the presence, the distribution or the clinical significance of Spn expression in human HCC. Therefore, we examined Spn expression in HCC tissue by immunohistochemistry and quantitative reverse transcriptase–PCR (RT–PCR). Correlation analyses of Spn expression and other possible interacting partners including microRNA-106a*, p53 and p14ARF were performed. Consequently, the role of Spn as a valuable novel prognostic marker was evaluated by correlating its expression level with several clinicopathological features as well as disease-free and cancer-specific survival in a large cohort of HCC patients. Finally, we explored the influence of reduced Spn expression on proliferation by correlating its expression with the Ki-67 proliferation marker in HCC tissue. Functional studies to explore the effects of reduced Spn expression on cellular growth were performed by small interfering RNA (siRNA)-mediated Spn silencing in cell line experiments.
Materials and methods
Patients and tissue samples
One hundred and four patients with primary HCC who underwent a curative liver resection at the Medical University of Graz (Austria) were included in this retrospective study. Tissue samples used in the study were retrieved from the Institute of Pathology, Medical University of Graz. We included consecutive patients diagnosed with HCC between January 1988 and December 2011. For an independent RT–PCR confirmation step, cryopreserved tissue of 10 HCC samples and their corresponding surrounding non-neoplastic liver were selected. The mean follow-up period was 3 years. None of the patients received neoadjuvant therapy or preoperative local treatment, and all underwent resection of the primary tumour. Postoperative surveillance was performed, including routine clinical and laboratory examinations every third month, and CT scans of the abdomen and X-rays of the chest every third month, respectively. After 5 years, the examination interval was extended to 12 months. The 7th edition of the American Joint Committee on Cancer (AJCC)/International Union Against Cancer (UICC) TNM system was used to classify the patients (Sobin et al, 2009). The study was approved by the local ethics committee of the Medical University of Graz (ID: 21–135 ex 09/10).
RNA isolation
For extraction of total RNA from cryopreserved liver tissue, 1 ml Trizol reagent (Invitrogen, Lofer, Austria) was added to a small piece of liver tissue (10 HCCs and their corresponding non-neoplastic liver tissue), which were already transferred into a tube filled up with MagNA Lyser Green beads (Roche Diagnostics, Vienna, Austria). Samples were homogenised by using MagnNA Lyser (Roche Diagnostics) at 6500 r.p.m. for 20 s, followed by incubation at room temperature for 5 min. Subsequently, 0.2 ml chloroform were added to each tube, agitated vigorously by hand for 15 s and incubated at room temperature for 3 min. Samples were centrifuged at 12 000 g at 4 °C for 25 min. The upper phase was transferred to a 1.5 ml eppendorf tube. Afterwards, 0.5-ml isopropanol were added, samples were vortexed for 15 s and incubated at room temperature for 10 min. Samples were centrifuged at 12 000 g at 4 °C for 10 min. Supernatants were removed and pellets were washed once with 1 ml of 75% ethanol. After centrifugation at 12 000 g at 4 °C for 10 min, supernatants were removed again and pellets were dried completely. Finally, pellets were dissolved in 30 μl Aqua bidest (Fresenius Kabi, Graz, Austria) and incubated at 56 °C for 10 min. RNA yield and quality were checked by using the NanoDrop 1000 Spectrophotometer (Peqlab, Erlangen, Germany). Total cellular RNA from HepG2 cells was isolated using the RNAeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Immunohistochemistry
Immunohistochemical analyses for Spn, p53, p14ARF and Ki-67 expression were performed on whole tissue slides of HCC tissue from all of the 104 patients. The 3-μm-thick sections were deparaffinised in xylene and rehydrated with graded ethanol. For Spn detection, the sections were subjected to antigen retrieval in a pressure cooker (Dako, Glostrup, Denmark; Pascal) in 0.01 M sodium-citrate buffer, pH 6.0, and subsequently incubated for 60 min with a rabbit antibody to human Spn (Millipore; anti-Spn antibody: AB5669 Vienna, Austria) at a 1 : 50 dilution. The reaction was visualised using the UltraVision LP Large Volume Detection System HRP Polymer (Thermo Scientific, Vienna, Austria), and all sections were counterstained with haematoxylin. For the negative control, the primary antibody was omitted. The frequency and the intensity of Spn-positive cells were estimated by two independent experienced observers (AA and MP) on a semiquantitative scoring system. The percentage of positive cells was rated as follows: 0, negative; 1, 1–10% positive cells; 2, 11–50% positive cells; 3, 51–80% positive cells; and 4, >80% positive cells. The staining intensity was scored as 0, negative; 1, weak; 2, moderate; and 3, intensive. The scores for the percentage of positive cells and the scores for the expression intensities were multiplied to calculate an immunoreactive score (IRS) (Remmele and Stegner, 1987). We separated the HCC patients into four groups according to their cytoplasmic expression of Spn (25th percentiles): group 1, IRS 0–2; group 2, IRS 3 and 4; group 3, IRS 6–8; and group 4, IRS 9–12.
For p53 staining, slides were incubated at 60 °C for 1 h and deparaffinised by xylene two times for 10 min. Sections were rehydrated by decreasing ethanol concentrations from 100–90% to 70% and to 50%, followed by a water bath for 40 min. Immunohistochemistry for p53 was performed on a DAKO autostainer (Dako), where a heat-induced epitope retrieval was performed and the antibody against p53 (monoclonal mouse anti-human p53 protein clone DO-7, M7001; Dako) was diluted 1 : 100. Dako Real (Dako; K5001) was used as a detection system. For visualisation, AEC Substrate Chromogen Ready-to-Use Kit (K3464; Dako) was utilised.
The percentages of tumour cells with a strong immunoreactive nuclear signal, indicating p53 accumulation in the nucleus, in the total number of tumour cells were counted in each of 10 high-power fields (magnification, × 40; 1 high-power field=0.24 mm2). The mean percentage of immunoreactive tumour cells was then semiquantitatively calculated on a 10-point scale (0–100%).
For p14ARF detection, antigens were unmasked by treating the slides with EDTA-sodium buffer (1 mM at pH=8.0) for 20 min in a microwave. After a cooling phase of 20 min, endogenous peroxidase was blocked with 3% H2O2 (Merck, Darmstadt, Germany) for 10 min at room temperature. A p14ARF antibody (GeneTex; GTX 23642, Irvine, CA, USA) was prepared in a 1 : 20 dilution in antibody diluent (Dako) and incubated for 60 min at room temperature. After washing and incubation with the biotinylated secondary antibody (K5001; Dako) for 30 min at room temperature, streptavidin-conjugated horse radish peroxidase (Dako) was added for additional 30 min at room temperature. AEC Substrate Chromogen Ready-to-Use (K3464; Dako) was used as the detection system for a minimum of 7 min. Colour development was monitored under a microscope. Sections were counterstained with Mayer’s hemalaun for 1 min.
The percentages of tumour cells with a strong immunoreactive cytoplasmic signal to p14ARF in the total number of tumour cells were counted in each of 10 high-power fields (magnification, × 40; 1 high-power field=0.24mm2). The mean percentage of immunoreactive tumour cells was then calculated and scored on a 10-point scale (0–100%).
The Ki-67 immunohistochemistry was analysed as a marker of proliferative activity in HCCs. For the detection of Ki-67, the anti-Ki-67 (30-9) antibody (Rabbit-a-confirm Ki-67 MIB Clone 30-9,Ready-to-Use; Ventana) was used on an autostainer (Ventana) by applying the automated Ventana staining system program CC1 mild (Ventana, Tucson, AZ, USA). The antibody was incubated for 32 min and for visualisation the iVIEW DAB Detection system (Ventana) was used. The number of positively stained nuclei of tumour cells was counted in at least 1000 tumour cells. The Ki-67 proliferation index was classified in accordance with the percentage of positive tumour cells as described previously (Kitamura et al, 2011).
Quantitative RT–PCR analysis of mRNA and miR-106a*
For mRNA quantification, up to 500 ng of total RNA was reverse transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. Quantitative RT–PCR was carried out in triplicates for each sample using commercially available primers specific for Spn (Hs_PPP1R9B_1_SG QuantiTect Primer Assay; Qiagen), the proliferation marker cyclin D2 (Hs_CCND2_1_SG QuantiTect Primer Assay; Qiagen) and the two housekeeping genes HMBS (Hs_HMBS_1_SG QuantiTect primer assay) and GAPDH (Hs_GAPDH_1_SG QuantiTect Primer Assay, Qiagen) on a LightCycler 480 Real-Time PCR System (Roche Diagnostics) using QuantiTect SYBR Green PCR Kit (Qiagen). Geometric mean of the housekeeping genes HMBS and GAPDH was used for normalisation, and relative gene expression levels were calculated with the delta delta CT method as previously described (Aigelsreiter et al, 2009). For detection of miR-106a* in cryopreserved HCC tissue samples, 500 ng of total RNA was reverse transcribed by the miScript II RT Kit (Qiagen) according to the manufacture’s protocol. The Hs_miR-106a*_1 miScript Primer Assay (mature microRNA (miRNA) sequence: 5′-CUGCAAUGUAAGCACUUCUUAC-3′ Qiagen), the Hs_miR-98_1 miScript Primer Assay (Qiagen) as a positive control (with previously described expression in liver tissue (Sukata et al, 2011)) and for normalisation of the Hs_RNU6-2_1 miScript Primer Assay (Qiagen) were applied on a LightCycler 480 Real-Time PCR System (Roche Diagnostics) using miScript SYBR Green PCR Kit (Qiagen).
Cell culture
The human hepatoma cell line HepG2 was purchased from American Type Culture Collection (Manassas, VA, USA). Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (1 × ) (+4.5 g l−1 D-glucose, L-glutamine, Pyruvat) (both form Gibco, Vienna, Austria), containing 10% foetal bovine serum gold (PAA Laboratories, Pasching, Austria) and antibiotics (penicillin and streptomycin) and were incubated in a 5% CO2 humidified incubator at 37 °C.
WST-1 proliferation assay
After standard trypsinisation, 10 000 HepG2 cells per well were seeded in 96-well culture plates and incubated in normal growth medium DMEM (Gibco) for 48 h after transfection of siRNA. The WST-1 proliferation reagent (Roche Applied Science, Vienna, Austria) was applied according to the manufacturer’s recommendations and after 4 h the colorimetric changes were measured using a SpectraMax Plus (Molecular Devices, Sunnyvale, CA, USA) at 450 nm with a reference wavelength at 620 nm. For each condition, cells were seeded in six-well plates and each experiment was independently repeated three times.
Transfection of Spn-targeting siRNA
For a transient transfection approach to reduce the Spn mRNA expression, HepG2 cells were transfected using the fast-forward transfection procedure as suggested by the HiPerFect Transfection Reagent (Qiagen) protocol according to the manufacturer’s instructions. A specific siRNA, which has been previously experimentally validated by the supplier to target the Spn mRNA (Hs_PPP1R9B_7 FlexiTube siRNA; Qiagen), was commercially purchased. For the reference control, we used the Allstars negative control siRNA (Qiagen), which is a validated siRNA without any functional consequence or known target sequence in human cells. For confirmation of transfections efficacy, the AllStars cell death control (Qiagen) were used within the same concentration as the target siRNA and reference control (20 nM). For siRNA delivery, 10 000 HepG2 cells in complete RPMI medium were seeded in 96-well plates and siRNA-HiPerFect Reagent transfection complexes were added according to the fast-forward transfection as recommended by the manufacturer (Qiagen). Transfected cells were incubated under their normal growth conditions (37 °C, 5% CO2) and the effect on Spn gene silencing was measured by quantitative RT–PCR after 48 h. Effects on cellular growth were monitored after 48 h by the WST-1 proliferation reagent assay and cyclin D2 expression was measured as described above.
Statistical analysis
All statistical analyses were performed using SPSS version 17.0 software (SPSS Inc., Chicago, IL, USA). Fisher’s exact test, the χ2-test, the Mann–Whitney U-test and the Student’s t-test procedure were used where appropriate to analyse Spn protein expression in relation to each clinicopathological parameter, cellular growth rates and gene expression. Spearman’s correlation was calculated to compare immunohistochemical Spn expression vs Spn mRNA expression, as well as p53, p14ARF and Ki-67 staining. Student’s t-test was used to test for differences in cellular growth rates. Disease-free survival (DFS) and cancer-specific survival (CSS) of the patients were calculated using the Kaplan–Meier method, and the differences were compared by the log-rank test. Backward stepwise multivariate Cox proportion analysis was performed to determine the influence of Spn expression, T classification, tumour grade, patient age and gender on DFS and CSS. The hazard ratios (HRs) estimated from the Cox models were reported as relative risks with corresponding 95% confidence intervals (CIs). A P-value of <0.05 was considered significant.
Results
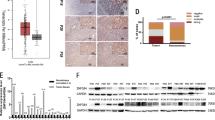
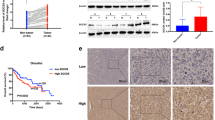
Clinicopathological patient characteristics, including gender, age, tumour stage, tumour grade, growth pattern and presence of liver cirrhosis, are detailed in Table 1. Tumour grades were G1 in 29 cases (27.9%), G2 in 60 cases (57.7%) and G3 in 15 cases (14.4%). Overall, there were more HCCs in male patients (76.9%), in cirrhotic livers (62.5%) and with trabecular growth pattern (87.4%). Regarding immunohistochemical detection of Spn, we found that in the non-neoplastic liver, Spn staining was predominantly cytoplasmic in a fine granular manner (Figures 1A and B). In non-cirrhotic livers, the positive staining was homogenous, whereas in cirrhotic nodules, the staining pattern was enhanced at the periphery of the nodules. In portal tracts, some inflammatory cells, such as lymphocytes and macrophages, showed positive Spn staining. The staining in the neoplastic liver was restricted to the cytoplasm and displayed a plump granular staining pattern compared with the fine granules that were predominantly found in the non-neoplastic liver (Figures 1C–E). Spinophilin staining was completely lost in 44 of 104 (42.3%) cases (Figures 1F–H) and reduced levels were found in an additional 37 (35.6%) cases. To confirm the reduced levels of Spn expression in HCC samples by an second independent method, we performed measurement of Spn expression by quantitative RT–PCR analysis on 10 HCC and 10 corresponding surrounding non-neoplastic liver samples. In 7 out of 10 cases (70%), a significantly reduced Spn mRNA expression in tumour tissue compared with the corresponding non-neoplastic tissue could be detected (P<0.05; Figure 2). There was also a significant correlation between Spn mRNA data and Spn immunohistochemistry on these cases (P=0.03, Spearman’s ρ). MiR-106a* expression, a microRNA that has been previously reported as a possible Spn-interacting partner in lung tumours (Molina-Pinelo et al, 2011), was not detected in any of these liver samples (data not shown). To analyse concurrent molecular alterations in other proteins, we performed immunohistochemical analyses of p53 and p14ARF in all 104 cases. Overall, there were 5 of 104 cases (5%) identified with positive nuclear p53 staining and 44 of 104 cases (33%) with a loss of p14ARF staining. There were no correlations found for the number of Spn-positive tumour cells and p53 (P=0.132, Spearman’s ρ) or p14ARF staining (P=0.847, Spearman’s ρ). To further evaluate whether Spn staining is associated with either clinicopathological parameters or the outcome of patients with HCC, we correlated immunohistochemical Spn staining results with tumour stage, tumour grade, the presence of cirrhosis, gender and age. We found no correlation between the level of Spn staining and the clinicopathological parameters analysed in this study (all P-values >0.05, data not shown). A 10-year DFS was observed in 21 (20.2%) patients, whereas disease recurred in 83 (79.8%) patients, and 76 (73.1%) patients died during the follow-up period. We divided the HCCs according to the semiquantitative IRS into four groups: no staining (n=75), weak (n=6), moderate (n=9) and strong (n=14). The interobserver disagreement between the two observers was low (κ-value=0.98). For the 10-year CSS, there was no significant difference between negative vs weak staining (P=0.426), there was a significant difference between weak vs moderate staining (P=0.041) and there was no significant difference between moderate and strong staining (P=0.832, pairwise log-rank tests). Therefore, we decided to dichotomise the HCC cases by assigning negative or weak staining samples to an Spn low expression group and moderate or strong staining samples to an Spn high expression group. Using this cutoff value for Spn staining, we found 81 (77.8%) of 104 HCC cases with Spn low expression and 23 (22.2%) of 104 cases with Spn high expression. Among the 104 HCC patients, disease recurrence was diagnosed after the 10-year follow-up in 69 of 81 (85.2%) cases with Spn low expression and in 14 of 23 (60.9%) cases with Spn high expression (P=0.01, log-rank test). For survival, death occurred in 62 of 81 (76.5%) cases with Spn low expression and in 13 of 23 (56.5%) cases with Spn high expression (P=0.006, log-rank test). Figures 3 and 4 show the Kaplan–Meier curves for the 10-year DFS and the 10-year CSS. These figures show that low Spn expression in neoplastic tissue is a factor for poor prognosis in HCC patients. Applying univariate Cox proportional analysis, low Spn expression (P=0.004) and a high T stage (P=0.022) were statistically significantly associated with a poor 10-year CSS, whereas no differences could be observed for the presence of cirrhosis (P=0.136), high tumour grade (P=0.121), age over 65 years (P=0.480) or gender (P=0.502). To determine the independent prognostic value of low Spn expression on 10-year DFS and CSS, a multivariate Cox proportional hazard analysis was performed, including age, gender and all parameters that were significantly associated with survival in the univariate analysis (Spn expression and T stage). Low Spn expression was also identified by multivariate analysis as an independent predictor for the 10-year DFS (HR=1.8, CI=1.04–3.40, P=0.043) and CSS in HCC (HR=2.0, CI=1.1–3.8, P=0.025). In an attempt to further clarify the association of impaired clinical outcome in HCC with reduced Spn expression, we correlated Spn immunohistochemical data vs the widely used proliferation marker Ki-67. The mean percentage of Ki-67-positive tumour cells in our cohort was 9.7±16.6%. A significant difference in Ki-67-positive cells between the Spn low compared with the Spn high expression group (11.5% vs 4.1% Ki-67-positive tumour cell, P=0.024, Mann–Whitney U-test) could be detected. Also, a significant inverse correlation between Spn expression and Ki-67 labelling exists (P=0.014, Spearman’s ρ). For further exploring the possible in vitro effects of reduced Spn levels in HCC cells, we established a transfection protocol for an Spn-targeting siRNA in HepG2 cells. After optimisation, a relative reduction of about 80% Spn mRNA expression could be reached (Supplementary Figure S1 and Supplementary Data section). Consecutively, the WST-1 assay was used to investigate the effect of Spn silencing on cellular growth of the transfected HepG2 cells. After 48 h, we observed a statistically significant increase in the growth of Spn-silenced cells in comparison to cells treated by the reference control Allstars negative control (30% increase, P<0.05; Figure 5). Finally, we measured cyclin D2 gene expression in Spn-silenced cells in comparison to cells treated by the reference control and detected a 50% higher cylin D2 expression level in Spn-silenced cells (P<0.05; Figure 6).
Representative micrographs of immunohistochemical staining of Spn in non-neoplastic and neoplastic liver tissue. (A and B) Non-neoplastic liver with a portal tract in the centre showing positive Spn staining in the surrounding hepatocytes. The staining pattern is fine granular and confined to the cytoplasm of the hepatocytes (magnification – A: × 20; B: × 40). (C) Non-neoplastic liver on the left side of the micrograph showing fine granular staining, whereas the corresponding HCC tissue on the right side displays a plumper granular fashion in the cytoplasm (magnification – × 20). (D and E) Hepatocellular carcinoma tissue showing positive plump granular cytoplasmic Spn staining (magnification – D: × 40; E: × 60). (F and G) Non-neoplastic liver tissue positive for Spn on the right side, whereas the HCC tissue on the left side is negative (magnification – F: × 20; G: × 40). (H) Hepatocellular carcinoma negative for SPN. Note single positive macrophages between the tumour cell trabeculae (magnification – × 60).
Expression analysis of Spn mRNA measured by RT–PCR in different human HCC samples (grey bars) and their corresponding non-neoplastic (black bars) liver tissue ( n =10). Seven out of 10 HCC samples showed a significantly reduced level of Spn mRNA when compared with the corresponding normal tissue (P<0.05).
Discussion
Spinophilin has been previously described as a cytoplasmic scaffold protein that interacts with a number of membranous and cytoskeletal proteins (Satoh et al, 1998). Because of its modular molecular structure, several interaction partners, including protein phosphatase 1 (PP1), F-actin, guanine nucleotide exchange factors, G-protein signalling protein regulators, membrane receptors, ion channels and seven-transmembrane receptors, have been identified (Carnero, 2012). Originally, Spn was discovered as an essential player involved in many physiological processes in the nervous system (Hsieh-Wilson et al, 1999; Yan et al, 1999; Allen et al, 2006). The first link between Spn and cancer was presented by a study by Vivo et al (2001), who discovered a role for Spn in the cell growth of mammalian cells (Vivo et al, 2001). They identified an interaction of Spn with the well-known tumour suppressor protein p14 alternate reading frame (p14ARF) (Vivo et al, 2001). More recently, studies have indicated additional roles for Spn in human cancers, including an inhibitory impact on the anchorage-independent growth of glioblastoma cells (Santra et al, 2006) and an influence on self-renewal and differentiation in brain tumour stem cells (Santra et al, 2011). Molina-Pinelo et al (2011) recently reported an absence or reduced levels of Spn in nearly 60% of human lung tumours and found that the decreased Spn levels correlated with a more malignant tumour grade.
In this study, we provide the first analysis of the putative tumour suppressor Spn in human HCC tissue and its association with patient clinicopathological data and clinical outcome. According to our data, which are in line with the reduced levels of Spn found in lung and other cancer types (Molina-Pinelo et al, 2011; Carnero, 2012), Spn immunoreactivity was significantly reduced in a substantial proportion of HCC cases compared with their surrounding non-neoplastic liver tissue. These findings were endorsed by an independent confirmation step by quantitative RT–PCR on a smaller set of samples, which significantly correlated with data from immunohistochemistry. In general, several molecular mechanisms for the loss of Spn protein in human HCCs might be considered. Rearrangement on chromosome 17q is a feature commonly found in human HCCs and could be the principal cause of the loss of Spn expression (Furge et al, 2005). It may be hypothesised that DNA hypermethylation, the occurrence of mutations in the Spn gene or Spn-targeting miRNAs lead to a loss of Spn expression in HCC cells. Although miR-106a* has been reported as a possible regulating factor of Spn expression in lung cancer cells (Molina-Pinelo et al, 2011), we lacked to detect this miRNA in liver tissue and could therefore not establish a role for miR-106a* in Spn silencing or hepatocarcinogenesis. In addition to the frequently reduced Spn levels in HCC, our investigations revealed that low Spn expression is associated with a lower DFS and a lower 10-year survival rate in patients with HCC. Importantly, our study revealed that low Spn expression independently predicts a higher risk of disease relapse and death after multivariate adjustment for other prognostic factors. To the best of our knowledge, no studies have addressed the topic of Spn expression in human HCC thus far. Given the lack of experimental data that could explain the association between the reduced Spn level and poor clinical outcome in HCC patients, the underlying mechanisms remain speculative. One possible explanation might be the previously published link to the p14ARF tumour suppressor protein (Vivo et al, 2001). p14ARF acts as an upstream regulator of the retinoblastoma (Rb)-cyclin dependent kinase-4 and p53 pathways, thereby inducing cell cycle arrest (Vivo et al, 2001). Previously published studies reported frequent inactivation of p14ARF in approximately 15–40% of HCCs (Tannapfel et al, 2001; Peng et al, 2002; Anzola et al, 2004), which is corroborated by our study as we found a loss of p14ARF in about 30% of HCCs. A diminished function or loss of p14AR has been shown to promote the invasion and migration of hepatocellular cancer cells (Chen et al, 2008), and the loss of p14ARF is associated with higher telomerase activity in HCC (Zhang et al, 2008). Thus, impairment of the Spn–p14ARF interaction in HCCs caused by decreased Spn expression might influence p14ARF-mediated tumour-suppressive functions. Another pathophysiological factor might be the interaction between Spn and PP1. Spinophilin regulates PP1 catalytic subunit activity (MacMillan et al, 1999), and the loss of Spn is associated with low PP1 phosphatase activity (Ferrer et al, 2011a). Protein phosphatase 1 is one of the key eukaryotic serine/threonine protein phosphatases, with significantly elevated activity in human HepG2 and rat AH13 hepatoma cells compared with primary cultured hepatocytes (Imai et al, 1999). The PP1 inhibitor microcystin-LR selectively induces liver damage and promotes hepatocarcinogenesis (Komatsu et al, 2007). Protein phosphatase 1 inhibition in the liver cancer cell line Huh-7 modulates oncogenic RNA alternative splicing to devitalise the cancer cells (Chang et al, 2011). Taken together, reduced expression of Spn might lead to altered PP1 activities that, in turn, would affect PP1-mediated functions in HCC. Interestingly, PP1 is also involved in the mitotic dephosphorylation of phosphorylated Rb protein (Tamrakar et al, 2000), as well as in the dephosphorylation of specific residues of p53 (Rubin et al, 1998), thereby regulating the control of cell cycle progression (Rubin et al, 1998). A recently published study by Ferrer et al (2011b) showed that the loss of Spn reduced PP1 activity, resulting in a high level of proproliferative phosphorylated Rb, which in turn leads to a compensatory increase in p14ARF and p53 activity. However, in the absence of p53 or mutated p53, reduced levels of Spn enhanced the tumorigenic potential of the cells, whereas ectopic overexpression of Spn in these human tumour cells greatly reduced cell growth (Ferrer et al, 2011b). Because the nuclear accumulation of p53 largely correlates with mutated p53, we tested whether the loss of Spn staining is correlated with nuclear p53 staining but found no significant correlation between Spn and p53 or p14ARF immunohistochemistry. Overall, we found rather low numbers of potential p53-mutated HCC cases in our cohort (5%). As shown previously, the incidence of p53 gene abnormalities in HCC varies in different geographical areas, which is due to different underlying environmental and genetic risk factors, resulting in less frequently encountered p53 mutations in Europe (Boix-Ferrero et al, 1999). Although the mechanisms discussed above could be considered as potentially involved in HCC, we could not establish any correlation between p14ARF or p53 expression and Spn immunoreactivity. Thus, other factors might also be relevant for the pathophysiolocial role of reduced levels of Spn in tumour progression of HCC. However, these factors need further experimental and clinical validation before a p53-independent tumorigenic role of Spn in HCC could be broadly accepted. Recently, Carnero (2012) reported that Spn knockout mice had decreased lifespan with increased cellular proliferation in different tissues. To explore a possible pathophysiological role of reduced Spn expression in HCC, we measured the widely used proliferation marker Ki-67 on the whole cohort to determine the proliferative activity (Kitamura et al, 2011). Overall, we detected a significant inverse correlation between Spn expression and proliferative activity in HCC. Therefore, we established an siRNA-based Spn silencing approach to study the effects of reduced Spn levels in tumour cells. The transfected cells showed a significant increase of cellular growth and an increased expression of the proliferation marker cyclin D2 when Spn mRNA was silenced. These in vivo and in vitro data support the hypothesis that a reduced Spn expression might act as a growth-promoting factor in HCC.
In summary, we identified that a substantial number of human HCCs show reduced or absent Spn immunoreactivity and mRNA expression, a finding that is compatible with the previously observed loss of Spn in lung adenocarcinoma and other types of cancer. The low expression of Spn in tumour tissue is an independent negative prognostic factor for clinical outcome in HCC patients. Spinophilin expression inversely correlates with proliferative activity in vivo and cellular growth in vitro. Further preclinical studies are warranted for the validation of Spn as a novel prognostic biomarker in HCC.
Change history
14 May 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Aigelsreiter A, Haybaeck J, Schauer S, Kiesslich T, Bettermann K, Griessbacher A, Stojakovic T, Bauernhofer T, Samonigg H, Kornprat P, Lackner C, Pichler M (2012) NEMO expression in human hepatocellular carcinoma and its association with clinical outcome. Hum Pathol 43 (7): 1012–1019
Aigelsreiter A, Janig E, Sostaric J, Pichler M, Unterthor D, Halasz J, Lackner C, Zatloukal K, Denk H (2009) Clusterin expression in cholestasis, hepatocellular carcinoma and liver fibrosis. Histopathology 54 (5): 561–570
Allen PB, Zachariou V, Svenningsson P, Lepore AC, Centonze D, Costa C, Rossi S, Bender G, Chen G, Feng J, Snyder GL, Bernardi G, Nestler EJ, Yan Z, Calabresi P, Greengard P (2006) Distinct roles for spinophilin and neurabin in dopamine-mediated plasticity. Neuroscience 140 (3): 897–911
Anzola M, Cuevas N, Lopez-Martinez M, Martinez de Pancorbo M, Burgos JJ (2004) p16INK4A gene alterations are not a prognostic indicator for survival in patients with hepatocellular carcinoma undergoing curative hepatectomy. J Gastroenterol Hepatol 19 (4): 397–405
Boix-Ferrero J, Pellin A, Blesa R, Adrados M, Llombart-Bosch A (1999) Absence of p53 gene mutations in hepatocarcinomas from a Mediterranean area of Spain. A study of 129 archival tumour samples. Virch Archiv Int J Pathol 434 (6): 497–501
Carnero A (2012) Spinophilin: a new tumor suppressor at 17q21. Curr Mol Med 12 (5): 528–535
Chang JG, Yang DM, Chang WH, Chow LP, Chan WL, Lin HH, Huang HD, Chang YS, Hung CH, Yang WK (2011) Small molecule amiloride modulates oncogenic RNA alternative splicing to devitalize human cancer cells. PLoS ONE 6 (6): e18643
Chen YW, Paliwal S, Draheim K, Grossman SR, Lewis BC (2008) P19Arf inhibits the invasion of hepatocellular carcinoma cells by binding to C-terminal binding protein. Cancer Res 68 (2): 476–482
Ferrer I, Blanco-Aparicio C, Peregrina S, Canamero M, Fominaya J, Cecilia Y, Lleonart M, Hernandez-Losa J, Ramon y Cajal S, Carnero A (2011a) Spinophilin acts as a tumor suppressor by regulating Rb phosphorylation. Cell Cycle 10 (16): 2751–2762
Ferrer I, Peregrino S, Canamero M, Cecilia Y, Blanco-Aparicio C, Carnero A (2011b) Spinophilin loss contributes to tumorigenesis in vivo. Cell Cycle 10 (12): 1948–1955
Frau M, Biasi F, Feo F, Pascale RM (2010) Prognostic markers and putative therapeutic targets for hepatocellular carcinoma. Mol Aspects Med 31 (2): 179–193
Furge KA, Dykema KJ, Ho C, Chen X (2005) Comparison of array-based comparative genomic hybridization with gene expression-based regional expression biases to identify genetic abnormalities in hepatocellular carcinoma. BMC Genom 6: 67
Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, Zucman-Rossi J (2012) Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 44 (6): 694–698
Hsieh-Wilson LC, Allen PB, Watanabe T, Nairn AC, Greengard P (1999) Characterization of the neuronal targeting protein spinophilin and its interactions with protein phosphatase-1. Biochemistry 38 (14): 4365–4373
Imai Y, Kakinoki Y, Takizawa N, Nakamura K, Shima H, Kikuchi K (1999) Up-regulation of nuclear PP1alpha and PP1delta in hepatoma cells. Int J Oncol 14 (1): 121–126
Kitamura K, Hatano E, Higashi T, Narita M, Seo S, Nakamoto Y, Yamanaka K, Nagata H, Taura K, Yasuchika K, Nitta T, Uemoto S (2011) Proliferative activity in hepatocellular carcinoma is closely correlated with glucose metabolism but not angiogenesis. J Hepatol 55 (4): 846–857
Komatsu M, Furukawa T, Ikeda R, Takumi S, Nong Q, Aoyama K, Akiyama S, Keppler D, Takeuchi T (2007) Involvement of mitogen-activated protein kinase signaling pathways in microcystin-LR-induced apoptosis after its selective uptake mediated by OATP1B1 and OATP1B3. Toxicol Sci 97 (2): 407–416
Lai Q, Avolio AW, Lerut J, Singh G, Chan SC, Berloco PB, Tisone G, Agnes S, Chok KS, Sharr W, Rossi M, Manzia TM, Lo CM (2012) Recurrence of hepatocellular cancer after liver transplantation: the role of primary resection and salvage transplantation in East and West. J Hepatol 57: 974–979
Lauwers GY, Terris B, Balis UJ, Batts KP, Regimbeau JM, Chang Y, Graeme-Cook F, Yamabe H, Ikai I, Cleary KR, Fujita S, Flejou JF, Zukerberg LR, Nagorney DM, Belghiti J, Yamaoka Y, Vauthey JN (2002) Prognostic histologic indicators of curatively resected hepatocellular carcinomas: a multi-institutional analysis of 425 patients with definition of a histologic prognostic index. Am J Surg Pathol 26 (1): 25–34
MacMillan LB, Bass MA, Cheng N, Howard EF, Tamura M, Strack S, Wadzinski BE, Colbran RJ (1999) Brain actin-associated protein phosphatase 1 holoenzymes containing spinophilin, neurabin, and selected catalytic subunit isoforms. J Biol Chem 274 (50): 35845–35854
Minguez B, Lachenmayer A (2011) Diagnostic and prognostic molecular markers in hepatocellular carcinoma. Dis Markers 31 (3): 181–190
Molina-Pinelo S, Ferrer I, Blanco-Aparicio C, Peregrino S, Pastor MD, Alvarez-Vega J, Suarez R, Verge M, Marin JJ, Hernandez-Losa J, Ramon y Cajal S, Paz-Ares L, Carnero A (2011) Down-regulation of spinophilin in lung tumours contributes to tumourigenesis. J Pathol 225 (1): 73–82
Peng CY, Chen TC, Hung SP, Chen MF, Yeh CT, Tsai SL, Chu CM, Liaw YF (2002) Genetic alterations of INK4alpha/ARF locus and p53 in human hepatocellular carcinoma. Anticancer Res 22 (2B): 1265–1271
Pons F, Varela M, Llovet JM (2005) Staging systems in hepatocellular carcinoma. HPB (Oxford) 7 (1): 35–41
Remmele W, Stegner HE (1987) Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Der Pathol 8 (3): 138–140
Rubin E, Tamrakar S, Ludlow JW (1998) Protein phosphatase type 1, the product of the retinoblastoma susceptibility gene, and cell cycle control. Front Biosci 3: D1209–D1219
Santra M, Santra S, Buller B, Santra K, Nallani A, Chopp M (2011) Effect of doublecortin on self-renewal and differentiation in brain tumor stem cells. Cancer Sci 102 (7): 1350–1357
Santra M, Zhang X, Santra S, Jiang F, Chopp M (2006) Ectopic doublecortin gene expression suppresses the malignant phenotype in glioblastoma cells. Cancer Res 66 (24): 11726–11735
Satoh A, Nakanishi H, Obaishi H, Wada M, Takahashi K, Satoh K, Hirao K, Nishioka H, Hata Y, Mizoguchi A, Takai Y (1998) Neurabin-II/spinophilin. An actin filament-binding protein with one pdz domain localized at cadherin-based cell-cell adhesion sites. J Biol Chem 273 (6): 3470–3475
Shariff MI, Cox IJ, Gomaa AI, Khan SA, Gedroyc W, Taylor-Robinson SD (2009) Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Exp Rev Gastroenterol Hepatol 3 (4): 353–367
Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62 (1): 10–29
Sobin LH, Gospodagorawicz MK, Wittekind C (2009) TNM Classification of Malignant Tumors 7th edn
Sukata T, Sumida K, Kushida M, Ogata K, Miyata K, Yabushita S, Uwagawa S (2011) Circulating microRNAs, possible indicators of progress of rat hepatocarcinogenesis from early stages. Toxicol Lett 200 (1–2): 46–52
Tamrakar S, Rubin E, Ludlow JW (2000) Role of pRB dephosphorylation in cell cycle regulation. Front Biosci 5: D121–D137
Tannapfel A, Busse C, Weinans L, Benicke M, Katalinic A, Geissler F, Hauss J, Wittekind C (2001) INK4a-ARF alterations and p53 mutations in hepatocellular carcinomas. Oncogene 20 (48): 7104–7109
Vivo M, Calogero RA, Sansone F, Calabro V, Parisi T, Borrelli L, Saviozzi S, La Mantia G (2001) The human tumor suppressor arf interacts with spinophilin/neurabin II, a type 1 protein-phosphatase-binding protein. J Biol Chem 276 (17): 14161–14169
Yan Z, Hsieh-Wilson L, Feng J, Tomizawa K, Allen PB, Fienberg AA, Nairn AC, Greengard P (1999) Protein phosphatase 1 modulation of neostriatal AMPA channels: regulation by DARPP-32 and spinophilin. Nat Neurosci 2 (1): 13–17
Zhang C, Guo X, Zhang L, Lu Z, Ma N, Cheng Y, Shen F, Zhang B, Wu M, Wei L (2008) Methylation-related silencing of p14ARF gene correlates with telomerase activity and mRNA expression of human telomerase reverse transcriptase in hepatocellular carcinoma. J Surg Oncol 98 (6): 462–468
Acknowledgements
This study was supported by grants from the Science Funds of the Styrian Government (to AA and MP), of the Hans und Blanca Moser Foundation (to FE) and from the START Research Initiative of the Medical University of Graz (to MP). The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115234, resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007–2013) and EFPIA companies’ in kind contribution (to JH).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Aigelsreiter, A., Ress, A., Bettermann, K. et al. Low expression of the putative tumour suppressor spinophilin is associated with higher proliferative activity and poor prognosis in patients with hepatocellular carcinoma. Br J Cancer 108, 1830–1837 (2013). https://doi.org/10.1038/bjc.2013.165
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.165