ABSTRACT
Environmental control of the alcohol dehydrogenase (Adh) and other stress response genes in plants is in part brought about by transcriptional regulation involving the G-box cis-acting DNA element and bZIP G-box Binding Factors (GBFs). The mechanisms of GBF regulation and requirements for additional factors in this control process are not well understood. In an effort to identify potential GBF binding and control partners, maize GBF1 was used as bait in a yeast two-hybrid screen of an A. thaliana cDNA library. GBF Interacting Protein 1 (GIP1) arose from the screen as a 496 amino acid protein with a predicted molecular weight of 53,748 kDa that strongly interacts with GBFs. Northern analysis of A. thaliana tissue suggests a 1.8-1.9 kb GIP1 transcript, predominantly in roots. Immunolocalization studies indicate that GIP1 protein is mainly localized to the nucleus. In vitro electrophoretic mobility shift assays using an Adh G-box DNA probe and recombinant A. thaliana GBF3 or maize GBF1, showed that the presence of GIP1 resulted in a tenfold increase in GBF DNA binding activity without altering the migration, suggesting a transient association between GIP1 and GBF. Addition of GIP1 to intentionally aggregated GBF converted GBF to lower molecular weight macromolecular complexes and GIP1 also refolded denatured rhodanese in the absence of ATP. These data suggest GIP1 functions to enhance GBF DNA binding activity by acting as a potent nuclear chaperone or crowbar, and potentially regulates the multimeric state of GBFs, thereby contributing to bZIP-mediated gene regulation.
Similar content being viewed by others
INTRODUCTION
Transcription factors are responsible for modulating gene expression via differential interaction with upstream DNA promoter sequences of targeted genes. The binding of transcription factors to regulatory elements then imparts a direct effect upon transcription. Therefore, these DNA binding proteins serve to provide essential termini for cellular signaling cascades that allow a cell to adjust to changing developmental or environmental conditions. In this critical role, the control of the DNA binding activity of transcription factors provides a direct means for cellular signals to be transduced into regulated gene activity.
There are several different classes of transcription factors, each recognizing a specific binding sequence. The sequences are not limited to a specific promoter, but rather can be present in several different genes thereby allowing for a single factor or group of related factors to affect the expression of a suite of proteins. One class of transcription factors, whose recognition sequence (CANNTG) is found in a variety of gene promoters, is the basic leucine zipper (bZIP) protein 1. bZIP proteins generally have a modular architecture that includes the basic amino acid region and the leucine zipper. The leucine zipper is a protein-interaction domain that allows for the formation of homo- and or heterodimers. The basic region is a positively charged helix that is responsible for contacting and binding DNA.
There are several recognized mechanisms for altering the apparent affinity of a bZIP protein for it's target DNA, most requiring direct interaction of the bZIP with cellular or nuclear factors 2. Phosphorylation of bZIP proteins by protein kinases can increase the specific binding of bZIP proteins to their target sequences 3, 4. Localization of bZIP proteins to areas outside the nucleus can result in inactivation by physically preventing DNA binding. Upon provision of the correct stimulus, the bZIP protein can be translocated to the nucleus and thereby made active 5. Dimerization adds another level of control by allowing the formation of homodimers or diverse heterodimers that can alter target specificity or affinity 1, 6. Recently, accessory proteins have been identified that can structurally modulate bZIP proteins in order to create modified DNA binding complexes 7. Many of these accessory proteins appear to have chaperone-like activities that result in increased DNA binding potential by the bZIP proteins.
The G-box Binding factors (GBFs) are a nearly ubiquitous family of bZIP proteins found in plants. GBFs bind to G-boxes, which are cis-elements containing dyad versions of the bZIP core sequence CCACGTGG. G-boxes are found within the promoters of several environmentally regulated plant genes and have been implicated as key elements in responses to cold, salt, light, dehydration, hypoxia and abscisic acid 8, 9.
There are specific examples of modifications to plant GBFs that result in changes to their effectiveness as transcription factors. For example, the binding of A. thaliana GBF1 can be enhanced by phosphorylation 10, while the activity of A. thaliana GBF2 is controlled by modifying the cellular localization of the protein upon blue light stimulation 5. Given the possibility of uncharacterized GBF family members and the propensity of bZIP proteins to undergo modifications, it is likely that there are undiscovered proteins that can also affect the intrinsic DNA binding properties of GBFs.
In an effort to identify accessory proteins that associate with plant GBFs, we have used maize GBF1 as bait in a yeast two-hybrid protein:protein screen. We report here the identification of G-box Interacting Protein 1 (GIP1). GIP1 binds transiently to both A. thaliana GBF3 and maize GBF1 and greatly enhances DNA binding in vitro and refolds denatured rhodanese, all of which suggest that GIP1 functions as a nuclear chaperone.
MATERIALS AND METHODS
Yeast two-hybrid screening
The coding region for maize GBF1 11 was fused to the C-terminal GAL4 DNA binding domain of the yeast two-hybrid binding vector pGBT9. The vector, pET15b GBF1, was used as a template in a PCR with the primers 5′-GAG GAT CCA GGC TCA GGA TGA-3′ and 3′-GCT AGT TAT TGC TCA GCG G-5′ to generate a BamHI GBF1 cassette. The cassette was subcloned into pGBT9 and sequenced to confirm orientation. pGBT9-GBF1 was transformed into competent HF7c yeast using the lithium acetate-based protocol described by the manufacturer (Clontech). Self-activation studies and protein extraction experiments were also performed as per the manufacturer's instructions. Western analysis of yeast extracts with anti-GBF1 antibodies was performed as described previously 11. All procedures used for library screening with the A. thaliana Matchmaker cDNA library (Clontech) were done in accordance with the manufacturers' protocols. Saccharomyces cerevisiae strain HF7c was cotransformed with pGBT9-GBF1 and the pGAD10 cDNA library. Positive clones that grew on –Trp/-Leu/-His media were screened for β-galactosidase (β-gal) activity by filter lifts to confirm protein-protein interactions using both reporter genes. Positive pGAD10-GIP clones were transferred to E.coli HB101 strain and subsequently re-cotransformed along with pGBT9-GBF1 or pTD1 (pGAD424-SV40 Large T-antigen) or pGAD424 (GAL4 Activation Domain) into Saccharomyces cerevisiae strain SFY526. Liquid β-galactosidase assays were performed with the SFY526 transformed yeast to determine relative binding affinities for the protein-protein interactions as compared to the positive controls, pVA3 (pGBT9 p53) and pTD1 standards that were supplied with the kit. Yeast two-hybrid positive clones were sequenced using automated sequencing.
Recombinant protein expression
pGAD10 A. thaliana GIP1 was digested with EcoRI to release the GIP1 coding region, which was subcloned into pETH3c 12. To introduce an N-terminal HIS6 tag for simplified purification of recombinant GIP1 protein, the GIP1 coding region was shuttled from pETH3c to pET15b HIS-tag vector (Novagen) as a Nde I / Cla I cassette. The pET15b A. thaliana GIP1 vector was transformed into competent BL21 DE3 cells and selected on ampicillin 2×YT plates. Vector-harboring cells were grown in Terrific Broth to a density of 0.1 OD at 600 nm and then induced with 1 mM isopropyl-thio-β-D-galatopyranoside for 3 h prior to centrifugal harvesting. The pellet was resuspended in immobilized metal chromatography binding buffer, as per manufacturer's instruction (Novagen, WI). Recombinant protein was purified essentially as described 13 except all reactions were carried out at 4ºC. Nickel column purified protein was dialyzed against Phosphate Buffered Saline (PBS) pH 7.6 at 4ºC and concentrated with Centricon concentrators (Amicon). The protein was stored at 4ºC and was stable for up to one month.
The expression vector and methods for the production of maize GBF1 (ZmGBF1) were as previously described 11. The expression vectors and methods for the production of A. thaliana GBF3a (AtGBF3a) and GBF3b (AtGBF3b) were as previously described 13. AtGBF3b is a truncated version of AtGBF3a containing the basic region and only portions of the proline-rich domain and the leucine zipper 13.
Antibody production and immunolocalization
Rabbit polyclonal GIP1 and AtGBF3a antibodies were produced commercially (Bioworld) from purified, His-tagged recombinant proteins. The sera were screened by ELISA or Western analysis using the corresponding recombinant proteins as positive controls. The sera were also screened against His-tagged recombinant maize GF14 12 14 as a negative control.
Nuclear extracts were prepared from A. thaliana cell suspension cultures as described 14. 20 μg of protein extract was loaded on a SDS-12% polyacrylamide gel. The electrophoresed proteins were transferred to nitrocellulose and blocked overnight with Blotto 15. The membranes were incubated with diluted sera (1/2000) in 5% nonfat dry milk containing PBS. Immunoreactive bands were detected by chemiluminescence according to the manufacturer's instructions (Pierce) and recorded on film.
Electrophorectic mobility shift assays
Recombinant AtGBF3a, AtGBF3b or ZmGBF1 was incubated with labeled, double-stranded G-box probe (3 ng G-box, 3 μg tRNA) with and without GIP1 or BSA in 20 mM Hepes-KOH, pH 7.6, 0.1 mM EDTA, 75 mM KCl, 10% glycerol and 5 mM β-mercaptoethanol (NEBD) at room temperature for 5 to 10 min before loading onto 6% polyacrylamide gels in 0.5×TBE buffer 13. Gels were dried onto DE81 paper, autoradiographed and analyzed on a Molecular Dynamics phosphoimager. The concentrations of individual protein stocks used in the binding mixtures were determined by Bradford analysis. For kinetic analysis of the effects of GIP1 on GBF binding, a serial dilution series of the GBF was prepared, and DNA binding in the presence and absence of GIP1 or BSA was determined on the same gel in order to eliminate differences due to exposure, time or other conditions. To compare relative effects of BSA and GIP1 on GBF binding, serial dilutions of BSA and GIP1 were prepared and analyzed against a single concentration of GBF within a single gel.
Lambda phagemid cDNA library screening
A BamHI / EcoRI fragment was released from pGAD10 GIP1 vector by restriction digestion and random prime labeled for probing an A. thaliana cDNA library in order to obtain a full length GIP1 cDNA clone. The library was commercially custom made from one-week old A. thaliana cv Columbia seedlings using the phagemid TriplEx2™ cloning vector system (Clontech). A total of approximately 72,000 plaques were screened, with positive clones carried through three successive rounds of titration plating. Isolated positive clones were transduced into E. coli BM25.8, promoting Cre recombinase-mediated release and circularization into pTripLex–GIP. The vectors were sequenced using automated sequencing.
Sequence data deposition
The sequences reported in the article have been deposited in the Genbank database (GIP1 accession No. AF344829, GIP2 accession No. DQ067323)
Genomic analysis of A. thaliana GIP1
Using the cDNA sequence from the yeast two-hybrid positive clones, a BLAST search was performed online through the NCBI website 16, 17 alignments and other computational manipulations were made in the programs MacVector 6.0 (Oxford Molecular) and Vector NTI (InforMax).
Northern analysis
Total RNA was extracted from A. thaliana root and inflorescence tissue as described previously 18. Samples of 20 μg total RNA per lane were loaded on a 2 % agarose / 6% formaldehyde gel. Electrophoresed RNA was transferred to Hybond N+ (Amersham Pharmacia Biotech) and probed with a GIP1 BamHI / EcoRI fragment labeled by random priming. Hybridization was recorded on Amersham Hyperfilm MP film (Amersham Pharmacia Biotech).
Gel filtration analysis of AtGBF3 after GIP1 incubation
Aggregation of AtGBF3a was induced by concentrating purified recombinant AtGFB3a to amounts in excess of 0.3 mg/ml in PBS using Centricon concentrators (Amicon). Aggregation was monitored by light scattering at 320 nm. The aggregated protein was incubated with a 1:1 molar ratio of recombinant GIP1 for 10 min at 4ºC prior to loading on a Superdex-200 high resolution gel filtration HR 10/30 analytical column (Amersham Pharmacia Biotech) equilibrated with PBS. Proteins were eluted with PBS at flow rates of 0.5 ml/min and detected by absorbance at 280 nm. Eluent was fractionated into 0.25 ml fractions and analyzed by western analysis with anti-AtGBF3a antibodies. Recombinant AtGBF3a without GIP1 incubation was also analyzed for comparison AtGBF3a incubated with GIP1 aliquots.
Refolding assays
Refolding experiments using GIP1 and GroEL / GroES with denatured rhodanese were performed essentially as described by Horowitz 19. GIP1 and GroEL / ES were used at 0.1 mg/ml concentrations for the assays and a 3×enzyme mixture was added to the substrate.
RESULTS
A. thaliana GIP1 interacts with maize GBF1 in a yeast two-hybrid library screen
Full-length ZmGBF1 was used as the bait for a yeast two-hybrid screen searching for A. thaliana GBF-interacting proteins. While both AtGBF3 and ZmGBF1 are potentially involved in Adh regulation, within their respective plants, in our hands ZmGBF1 is a more soluble factor and therefore was chosen as the primary bait. ZmGBF1 bait constructs were tested for protein expression and self-activation prior to library screening. As a eukaryotic transcription factor, ZmGBF1 could behave as an activator in yeast or be toxic to the cells. Soluble ZmGBF1 was expressed from the bait construct in the transformed yeast (Fig. 1A) with no reporter gene activity above the GAL4 binding domain vector background activity. When the A. thaliana GAL4 activation domain-linked cDNA library was cotransformed with pGBT9-GBF1 into competent HF7c yeast and plated onto –Trp/-Leu/-His media, growth and β-gal staining identified two classes of A. thaliana genes interacting with ZmGBF1. One class, represented by two separate clones of GIP1 (GBF Interacting Protein 1), gave the highest reporter activity and was characterized further. The other class, GIP2, interacted less strongly and was represented by a single clone that was severely truncated as compared to Northern transcript size (data not shown), therefore the GBF-binding sequence was simply deposited into GenBank (accession No. DQ067323). The pGAD10-GIP1 plasmids were isolated from yeast and cotransformed into SFY526 along with pGBT9-ZmGBF1 or pGBT9-SV40 Large T-antigen or pGBT9 DNA binding domain alone to confirm the interaction using more sensitive liquid β-gal assays. β-gal activities for the pGAD10-GIP1s and pGBT9-ZmGBF1 were ∼60 % of the positive control plasmids included with the yeast two hybrid kit. GIP1 was also tested for two-hybrid interaction with AtGBF3. The combination of pGAD10-GIP1 and pGBT9-AtGBF3 produced β-gal activities ∼24% of the positive controls. To test if GIP1 itself interacted directly with the GAL4 binding domain, pGAD10-GIP1s were cotransformed with the GAL4 binding domain vector pGBT9 without the GBF insert. No β-gal activity above background was detected (Fig. 1B).
Yeast expressed GBF1 interacts with GIP1 protein in the two-hybrid system. (A) Yeast cells containing ZmGBF1 fused to the GAL4 binding domain produced soluble recombinant fusion protein that can be detected with ZmGBF1 antibodies using ELISA. (B) β-galactosidase (β-gal) activity was calculated for yeast containing ZmGBF1 and GIP1, a positive clone from the library screen. The reporter activity was plotted relative to the positive controls pVA3 and pTD1.
cDNA and genomic structure of A. thaliana GIP1
The two pGAD10-GIP1 clones were sequenced and determined to be redundant partial clones of 1126 and 685 bases, differing in the poly A sequence as well as with their overall length. BLAST analysis initially revealed homology to the A. thaliana genomic DNA chromosome 3 clones ATAC067753 and AP000375. Subsequent to the completion of the Arabidopsis Genomic Initiative sequencing project, the gene coding for GIP1 was assigned to the locus At3g13222. Genomic sequence for the A. thaliana chromosome I BAC F20N2 clone AC002328 also displayed similarity with GIP1 cDNA, however this clone was divergent, especially at the 5′ region suggesting that it belongs to a close, but separate, family member. An A. thaliana cDNA library was screened by hybridization using pGAD10-GIP1 as a probe to identify longer GIP1 cDNA clones. The longest cDNA clone identified was 1618 bp (Genbank accession No. AF344829). The complete sequence was identified as locus At3g1322 / MJG19.5 (Genbank accession No. 820513) on chromosome 3. The gene product NM_112163, contains a 1704 bp CDS that encodes a 567 aa protein (Genbank accession No. NP_187929) with predicted molecular weight of 61812 kDa.
GIP1 mRNA is present in A. thaliana root and inflorescence
Northern analysis of total RNA from A. thaliana roots and inflorescences revealed a single GIP1 transcript of approximately 1.8–1.9 kb. The relative abundance was marginally higher in roots than in the inflorescence (Fig. 2A).
GIP1 expression and immunolocalization. (A) Northern analysis. Total RNA from A. thaliana roots (lanes 1 and 3) and inflorescences (lanes 2 and 4) was isolated and electrophoresed on a 2% formaldehyde RNA gel and stained with ethidium bromide (lanes 1 and 2) or transferred to Hybond N+ membrane (lanes 3 and 4). The membrane was probed with a radiolabeled GIP1 probe derived from the yeast two-hybrid clone pGAD10-GIP1. (B) Bacterially expressed GIP1 encoded by the yeast two-hybrid truncated clone pGAD10-GIP1 was electrophoresed using SDS-PAGE and stained with Coomassie brilliant blue (lane 1) or immunoblotted and probed with antibodies to recombinant GIP1 (lane 2). Immunoblot analysis of A. thaliana cytoplasmic (lane 3) and nuclear (lane 4) fractions was also performed using the same antibodies.
Recombinant GIP1 protein and GIP1 antibody production
Expression of the GIP1 coding sequence from pGAD10-GIP1 in the bacterial vector pET15b resulted in the production of a protein with an apparent molecular weight of 55 kDa (Fig. 2B). This protein was missing amino acids from the NH2 terminus based on the prediction from the cDNA and genomic sequence. The expressed protein was very susceptible to bacterial proteolytic degradation when analyzed by SDS-PAGE. The protein was also labile to freezing and had a limited shelf life of approximately one month at 4ºC. The GIP1 protein in PBS had a maximum solubility of approximately 0.3 mg/ml. Purification of recombinant GIP1 using immobilized metal affinity chromatography allowed for production of homogenous protein at levels sufficient for rabbit antibody production. GIP1 sera cross-reacted with both denatured and native GIP1 as assayed by Western analysis (Fig. 2B) and ELISA (data not shown), respectively.
A. thaliana GIP1 protein is present in nuclei
To ascertain the expression characteristics of cellular GIP1 protein, antibodies to recombinant GIP1 were used in Western analysis of A. thaliana tissue fractions. Nuclear fractions contained the majority of a ∼63 kDa cross-reactive protein, although a small amount was present in cytoplasmic fractions (Fig. 2B). The cellular GIP1 protein is close to the presumed molecular weight of GIP1 and predictably larger than the truncated version expressed in E. coli.
EMSA DNA binding enhancement by GIP1 addition
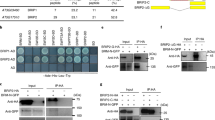
The presence of GIP1 increases the apparent affinity of GBFs for G-box DNA as determined by EMSA. In Fig. 3, the DNA binding capability of AtGBF3b is examined. AtGBF3b is a truncated version of AtGBF3 containing a very minimal leucine zipper, the basic region, and only part of the proline-rich domain 13. Over the range of concentrations from 0.01 to 0.000625 μg/μl, AtGBF3b demonstrates no DNA binding in EMSA. However, with the presence of 0.02 μg/μl GIP1, AtGBF3b demonstrated significant DNA binding that was limited to a clear, single species of protein-DNA complex.
Electrophoretic mobility gel shift analysis of recombinant A. thaliana GBF3b with GIP1. At GBF3b binds DNA in the presence of GIP1. In the absence of additional proteins, AtGBF3b was incapable of binding G-box DNA over the concentration range examined in this experiment (0.01-0.000625 μg/μl). However, addition of 0.02 μg/μl GIP1 enabled AtGBF3b to become a strong G-box binding protein.
This GIP1-dependent increase in DNA binding was not limited to AtGBF3b. GIP1 also increased the apparent DNA binding affinity of both AtGBF3a and ZmGBF1. AtGBF3a is a near full-length version of AtGBF3 containing the complete basic region and leucine zipper, as well as most of the proline-rich domain 13. In Fig. 4A, the kinetics of G-box binding by AtGBF3a is plotted against its concentration in the presence and absence of 0.02 μg/μl GIP1. For AtGBF3a, the apparent affinity for G-box DNA was increased by approximately 10-fold by the addition of GIP1 to the binding reaction. GIP1 had a similar effect on ZmGBF1, increasing the apparent affinity of ZmGBF1 by 10-fold as well (Fig. 4B).
GIP1 increases the DNA binding capacity of both AtGBF3a and ZmGBF1. The apparent DNA binding of AtGBF3a and ZmGBF1 were determined in the absence and presence of 0.02 μg/μl GIP1. The affinities of AtGBF3a (A) and ZmGBF1 (B) were each increased by approximately 10-fold, as determined by comparing the log [GBF] at 50% maximal binding in the presence and absence of GIP1. The DNA binding of ZmGBF1 was also tested in the presence of 15 μg/μl BSA. BSA also increased the apparent affinity of ZmGBF1, but to a lesser extent than GIP1.
Because earlier studies on ZmGBF1 and AtGBF3 binding activities were conducted with BSA present in the DNA binding mixtures, and because BSA is known to generally solubilize and stabilize protein structures, BSA was tested for its ability to enhance the DNA binding of GBFs. As shown in Fig. 4B, the presence of 15 μg/μl BSA did increase the apparent affinity of ZmGBF1 for G-box DNA, but to a lesser extent than the increase provided by GIP1. To compare directly the relative abilities of BSA and GIP1 to enhance DNA binding activity of GBFs, a dilution series of GIP1 and BSA were analyzed on the same EMSA gel using a constant concentration of GBF. As shown in Fig. 5, GIP1 was approximately 100-fold better than BSA at increasing the apparent DNA binding affinity of ZmGBF1, and >100-fold better than BSA at increasing the apparent affinity of AtGBF3a.
GIP1 is approximately greater than 100-fold more effective than BSA at increasing the DNA binding of GBF. At GBF3a and ZmGBF1 at 0.001 μg/μl were incubated with a serial dilution of GIP1 and a serial dilution of BSA then analyzed by EMSA. The counts bound by GBF are plotted against the concentration of effector (BSA or GIP1). At 50% maximal binding, GIP1 was approximately 100-fold better than BSA at enhancing ZmGBF1 binding, and >100-fold better at enhancing AtGBF3a binding.
In order to determine if GIP1 increases DNA binding by forming a stable complex with GBF, EMSA analysis was performed under conditions of protein concentrations that would highlight such complexes. In Fig. 6, the concentration of AtGBF3a (0.0015 μg/μl) was carefully chosen such that mobility of the AtGBF3a protein DNA complexes could be observed and compared in both the presence and absence of GIP1. A single protein DNA complex was observed for AtGBF3a in the absence of GIP1 (lane 1), and GIP1 alone showed no intrinsic DNA binding capability (lane 3). A mixture of the same amounts of AtGBF3a and GIP1 demonstrated an increase in DNA binding, but only a single protein DNA complex was observed, and the mobility of that complex was the same as that for AtGBF3a alone. There was no observable additional band that would indicate a GIP1:AtGBF3a:DNA supercomplex.
GIP1 does not form an EMSA-stable complex with GBF. EMSA analysis of AtGBF3a in the presence of GIP1 reveals a single protein:DNA complex (lane 1) that has the same electrophoretic mobility as AtGBF3a by itself (lane 2). GIP1 itself has no DNA binding activity (lane 3). The concentration of AtGBF3a (0.0015 μg/μl) was chosen to be high enough to allow DNA binding by AtGBF3a itself, as well as the enhancement of DNA binding activity provided by GIP1.
Chaperone activity and reduction of aggregation of GBFs by GIP1
To explore the possibility that GIP1 may enhance GBF3a DNA binding by chaperone activity or solubilzation, aggregated recombinant GBF3a was incubated with recombinant GIP1 and analyzed with FPLC. High resolution gel filtration chromatography of aggregated GBF3a alone identified complexes in the range > than 1.3×106 kDa (exclusion limit of the column) to 240 kDa (Fig. 7A). Addition of equimolar ratios of GIP1 prior to column separation of aggregated GBF3a resulted in a drastic reduction of the high molecular weight AtGBF3a complexes (1.3×106 to 240 kDa) and appearance of a range of species down to dimer and monomer-sized species. GIP1 was tested for general chaperone activity on the denatured enzyme rhodanese and found to possess chaperone activity in an ATP-independent manner (Fig. 7B).
GIP1 disaggregates GBF3a into lower molecular weight complexes including monomers and functions as a chaperone. (A) Equimolar ratios of aggregated GBF3a were incubated with recombinant GIP1 prior to loading on a Superdex-200 high resolution gel filtration column. Fractions were collected and analyzed with SDS-PAGE / immunoblot analysis with GBF3a antibody. As a control aggregated GBF3a without GIP1 addition was also analyzed using the same procedure. The shaded area beneath tracing represents redistribution of GBF3a after incubation with GIP1. (B) The ability of GIP1 to refold urea-denatured rhodanese was compared to GroEL / GroES with and without ATP. An aliquot of 0.1 mg/ml GIP1 (circle) or GroEL / ES (square) or buffer (star) was incubated with rhodanese in the presence of ATP. The same experiment was repeated in the absence of ATP, GIP1 (triangle), GroEL / GroES (diamond).
DISCUSSION
In an effort to identify how proteins interacting with bZIP proteins might assist in modulating transcription, we utilized the yeast two-hybrid system to screen for proteins that physically interact with ZmGBF1. One initial concern using this system was that, as a transcription factor, GBF fused to the GAL4 binding domain would self activate or be lethal to the yeast and therefore not function as useful bait. This was not the case, as the ZmGBF1 fusion protein was detectable in surviving yeast and only background reporter activity was observed in the absence of GAL4 activation domain-containing vectors. In library screening, ZmGBF1 interacted strongly with a novel protein, GIP1. Two variations of GIP1 clones (which differed by the length of the clone) were recovered; however both had the same level of interaction.
Amino acid sequence comparison of predicted GIP1 protein using Genbank database BLAST searches did not reveal any significant similarity to other proteins of known function. GIP1 does not contain a leucine zipper domain that would allow for interaction with GBFs as a heterodimeric partner, nor does it contain any motifs that that would lead to predictions of functional properties. Sequence analysis of the GIP1 CDS using InterProScan at the EMBL-EBI, did reveal a pfam06972: DUF1296 plant-specific motif of unknown function. BLAST analysis of GIP1 nucleic acid sequence identified two BAC genomic clones spanning the cDNA from our experiments. The At3g1322 gene organization is in good agreement with our cDNA data and further demonstrates the authenticity of GIP1. Furthermore, antibodies raised against GIP1 recognize a protein within A. thaliana nuclei that is consistent with the predicted size of GIP1, essentially confirming that GIP1 is a bona fide and expressed A. thaliana gene. Based upon BLAST results GIP1 appears to a plant specific protein with the closest homology occurring with rice.
The interaction between GIP1 and GBFs increased the apparent DNA binding capacity of GBFs, but without the formation of an overtly stable complex. In spite of the fact that GBF and GIP1 interact strongly in the yeast two-hybrid system, neither EMSA (Fig. 6) nor glycerol gradient analysis (data not shown) revealed any stable intermediate between them. EMSA clearly demonstrated that GIP1 did affect the ability of GBF3a to bind DNA, but without changing the gel mobility of the DNA / GBF3a complex. Since GIP1 by itself did not bind DNA, the role of GIP1 seems to be to modify the state of GBFs for enhanced DNA binding. While the addition of BSA could also help increase apparent DNA binding of GBFs by presumably increasing the solubility, the level of efficiency of BSA was <100 fold less than that of GIP1. Therefore GIP1 likely behaves as a specific nuclear-localized molecular chaperone.
Accessory proteins that can increase the binding potential of bZIP proteins have been identified in other eukaryotic systems. Two proteins, Tax from HTLV-I retrovirus and pX from the hepatitis B virus, contain possible metal binding regions that are thought to interact with the basic region to stimulate binding of bZIP proteins through enhanced bZIP dimerization. An additional feature of the pX-bZIP interaction is that the interaction can alter DNA recognition by pX 20. A human protein called bZIP-enhancing factor (BEF) also alters bZIP binding; however, BEF binds to the leucine zipper region 21. BEF's mode of regulation is apparently to recognize unfolded bZIP proteins and assist in proper folding for DNA binding. In A. thaliana, NPR1 enhances the DNA binding activity of the TGA family of bZIP transcription factors 22, 23, 24, 25. NPR1 is a central element of the salicyclic acid-mediated signal transduction pathway that regulates pathogen resistance. Interestingly, in a yeast two-hybrid screen, NPR1 interacts differentially with a subgroup of the TGA family allowing for specific signal transduction. The ability of NRP1 to function on other bZIP proteins, such as GBFs, is not known.
Although these accessory proteins may seem to be similar to GIP1 in ostensible function, they do not share sequence similarity with GIP1. Alignment between GIP1 and the other nuclear chaperones does not reveal any significant similarities. For example, the hallmark of NPR1 is the presence of ankyrin repeat domains, which mediate protein:protein interactions 26; however GIP1 does not have any putative repeats of this type. Tax and pX have apparent metal binding domains, which are absent in GIP1. In fact, there are no sequence domain homologies which suggest functional mechanisms for the GIP1 enhancement of GBF DNA binding activity. Therefore, GIP1 appears to be a new class of nuclear chaperone-like protein.
The mechanism(s) for nuclear chaperone activation of bZIP proteins appears to vary greatly, without a common theme. In addition, the identified domains that interact with bZIP proteins do not suggest a common mode of interaction. This complicates the ability to predict bZIP nuclear chaperones from consensus sequences and identify distinguishable enzymatic assays. Given that GIP1 appeared to solubilize active GBF3a, we examined if GIP1 could disassociate and solubilize intentionally aggregated, misfolded GBF3a. The result was a reduction in the molecular weights of the complexes GBF3a formed with itself. In the presence of GIP1, GBF3a was reduced through a range of sizes down to monomers, a state that normally represents a very small percentage of GBF3a species. The ability to efficiently form heterodimers from different bZIP proteins would seem to require the ability to separate bZIP proteins into monomer pools and allow for their association into dimers. An alternative premise is that bZIP proteins first bind DNA as monomers and then dimerize. This postulation also requires that bZIP protein dimers disassociate prior to DNA binding. Since GBFs exist preferentially as dimmers 27, a crowbar-like chaperone activity such as GIP1 would be required for efficient separation, dimerization and binding to the proper DNA target.
While understanding the complexities of the mechanism by which GIP1 activates GBFs is for future studies, it is clear that disassociation is part of the process. This dissociation mechanism has not been described for any other plant nuclear chaperones. It is unclear, for example, if NPR1 is capable of separation of bZIP dimers and functions in a similar manner. Additionally, since our two-hybrid screen for GBF binding partners produced two classes of GIPs (GIP1 and GIP2), other factors may function alone or in concert with GIP1 to provide selectivity or enhanced function. It seems likely that the delicate balance of regulatory transcription of environmentally induced genes using promiscuous factors such as GBFs is influenced by a host of auxiliary factors involved in the formation of protein DNA complexes. Certain helper proteins such as GIPs may be required for the attainment of an appropriately active transcriptional complex.
References
Hurst HC . Leucine zippers. Transcription Factors. San Diego: Academic Press, 1996.
Schwechheimer C, Zourelidou M, Bevan MW . Plant transcription factor studies. Annual Review of Plant Physiology and Plant Molecular Biology 1998; 49:127–150.
Meshi T, Moda I, Minami M, Okanami M, Iwabuchi M . Conserved Ser residues in the basic region of the bZIP-type transcription factor HBP-1a(17): importance in DNA binding and possible targets for phosphorylation. Plant Mol Biol 1998; 36:125–36.
Wellmer F, Kircher S, Rugner A, et al. Phosphorylation of the parsley bZIP transcription factor CPRF2 is regulated by light. J Biol Chem 1999; 274:29476–82.
Terzaghi WB, Bertekap RL Jr, Cashmore AR . Intracellular localization of GBF proteins and blue light-induced import of GBF2 fusion proteins into the nucleus of cultured Arabidopsis and soybean cells. Plant J 1997; 11:967–82.
Cheong YH, Yoo CM, Park JM, et al, Hong JC . STF1 is a novel TGACG-binding factor with a zinc-finger motif and a bZIP domain which heterodimerizes with GBF proteins. Plant J 1998; 15:199–209.
Baranger AM . Accessory factor-bzip-DNA interactions. Current Opinion in Chemical Biology 1998; 2:18–23.
Shinozaki K, Yamaguchi-Shinozaki K . Gene Expression and Signal Transduction in Water-Stress Response. Plant Physiol 1997; 115:327–34.
Menkens AE, Schindler U, Cashmore AR . The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci 1995; 20:506–10.
Klimczak LJ, Schindler U, Cashmore AR . DNA binding activity of the Arabidopsis G-box binding factor GBF1 is stimulated by phosphorylation by casein kinase II from broccoli. Plant Cell 1992; 4:87–98.
de Vetten NC, Ferl RJ . Characterization of a maize G-box binding factor that is induced by hypoxia. Plant J 1995; 7:589–601.
Hattori T, Vasil V, Rosenkrans L, et al. The Viviparous-1 gene and abscisic acid activate the C1 regulatory gene for anthocyanin biosynthesis during seed maturation in maize. Genes Dev 1992; 6:609–18.
Lu G, Paul AL, McCarty DR, Ferl RJ . Transcription factor veracity: is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell 1996; 8:847–57.
de Vetten NC, Ferl RJ . Transcriptional regulation of environmentally inducible genes in plants by an evolutionary conserved family of G-box binding factors. Int J Biochem 1994; 26:1055–68.
Harlow E, Lane D . Antibodies. Cold Springs Harbor: Cold Springs Harbor Laboratory, 1988.
Benson DA, Boguski MS, Lipman DJ, et al. GenBank. Nucleic Acids Res 1999; 27:12–7.
Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389–402.
Laughner BJ, Sehnke PC, Ferl RJ . A novel nuclear member of the thioredoxin superfamily. Plant Physiol 1998; 118:987–96.
Horowitz PM . Methods Mol Biol. In: Shirley BA, ed. Vol. 40. Humana Press: Totowa. 1995: 361–8.
Perini G, Oetjen E, Green MR . The hepatitis B pX protein promotes dimerization and DNA binding of cellular basic region/leucine zipper proteins by targeting the conserved basic region. J Biol Chem 1999; 274:13970–7.
Virbasius CM, Wagner S, Green MR . A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol Cell 1999; 4:219–28.
Zhou JM, Trifa Y, Silva H, et al. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact 2000; 13:191–202.
Niggeweg R, Thurow C, Weigel R, Pfitzner U, Gatz C . Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol Biol 2000; 42:775–88.
Despres C, DeLong C, Glaze S, Liu E, Fobert PR . The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 2000; 12:279–90.
Zhang Y, Fan W, Kinkema M, Li X, Dong X . Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci U S A 1999; 96:6523–8.
Cao H, Glazebrook J, Clarke JD, Volko S, Dong X . The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997; 88:57–63.
Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR . Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. Embo J 1992; 11:1261–73.
Acknowledgements
We would like to thank Dr. Curt HANNAH for use of the Pharmacia Superdex-200 column. We would also like to thank the ICBR DNA sequencing core for analysis of the yeast two-hybrid positive clones, the vector constructs and cDNA library clones. This research was supported by the U S Department of Agriculture Grants 00-35304-9601 and 98-35301-6083. This article is Florida Agricultural Experiment Station journal series number R-08010.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
SEHNKE, P., LAUGHNER, B., LINEBARGER, C. et al. Identification and characterization of GIP1, an Arabidopsis thaliana protein that enhances the DNA binding affinity and reduces the oligomeric state of G-box binding factors. Cell Res 15, 567–575 (2005). https://doi.org/10.1038/sj.cr.7290326
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290326
Keywords
This article is cited by
-
The lineage and diversity of putative amino acid sensor ACR proteins in plants
Amino Acids (2020)
-
GIP1 protein is a novel cofactor that regulates DNA-binding affinity of redox-regulated members of bZIP transcription factors involved in the early stages of Arabidopsis development
Protoplasma (2015)
-
TF-Cluster: A pipeline for identifying functionally coordinated transcription factors via network decomposition of the shared coexpression connectivity matrix (SCCM)
BMC Systems Biology (2011)