Abstract
SAG21/LEA5 is an unusual late embryogenesis abundant protein in Arabidopsis thaliana, that is primarily mitochondrially located and may be important in regulating translation in both chloroplasts and mitochondria. SAG21 expression is regulated by a plethora of abiotic and biotic stresses and plant growth regulators indicating a complex regulatory network. To identify key transcription factors regulating SAG21 expression, yeast-1-hybrid screens were used to identify transcription factors that bind the 1685 bp upstream of the SAG21 translational start site. Thirty-three transcription factors from nine different families bound to the SAG21 promoter, including members of the ERF, WRKY and NAC families. Key binding sites for both NAC and WRKY transcription factors were tested through site directed mutagenesis indicating the presence of cryptic binding sites for both these transcription factor families. Co-expression in protoplasts confirmed the activation of SAG21 by WRKY63/ABO3, and SAG21 upregulation elicited by oligogalacturonide elicitors was partially dependent on WRKY63, indicating its role in SAG21 pathogen responses. SAG21 upregulation by ethylene was abolished in the erf1 mutant, while wound-induced SAG21 expression was abolished in anac71 mutants, indicating SAG21 expression can be regulated by several distinct transcription factors depending on the stress condition.
Similar content being viewed by others
Introduction
In their natural environment, plants are subjected to a constantly fluctuating ensemble of abiotic and biotic stressors including rapid or prolonged changes in temperature, water availability, mechanical stress, e.g., wounding, and salinity, as well as attack from herbivores and pathogens. Stress-responsive signalling pathways enable plants to perceive these adverse environmental cues and transmit this information through a network of specific and common signals to help cells withstand short term or longer-term stresses1. This signalling induces changes at the molecular (transcriptomic and proteomic), cellular and physiological levels which can be persistent or short lived2. Signalling cascades are functionally related, but stressor specific, with responses to multiple stresses differing from responses to individual stresses3. However, many different abiotic stresses lead to imbalances in metabolic pathways which induce oxidative stress4, caused by the production and accumulation of reactive oxygen species (ROS). This can lead to changes in cellular redox status, direct oxidative modification of components within signalling pathways and oxidative damage to membrane proteins.
Transcriptional activation and repression occur through the binding of transcription factors (TFs) to sequence specific cis-elements within the target gene promoter. A total of 2296 TF genes have been identified within the Arabidopsis genome and classified into 58 families based on their DNA binding domains (Plant RegMap5). Of these 58 families, several are particularly involved in stress responses. These include the 71 WRKYs6,7, 117 NACs8, 122 ERFs9, and the 126 MYB family TFs10. The cis-elements to which these TFs bind are mostly well-defined, for example W-boxes in the case of WRKY TFs11, which share the core sequence TGAC, and the NAC recognition sequence (NACRS) for NAC TFs, although atypical cis-elements are also found12. Typically, ERF/AP2 family TFs bind to DRE/CRT promoter elements to activate abiotic stress responses and GCC-boxes to regulate biotic stress resistance9 although again divergent ERF TF binding motifs are also found.
Many WRKY TFs are part of biotic and abiotic stress signalling networks. At least eight Arabidopsis WRKY TFs are upregulated by reactive oxygen species6, and at least another six Arabidopsis WRKY TFs are activated by drought stress13. Interestingly, two WRKY TF genes, AtWRKY40 and AtWRKY63 regulate stress-responsive mitochondrial genes14 but not mitochondrial genes that are constitutively expressed. WRKY TFs are divided into three phylogenetically related groups (I–III) and group III is further divided into IIIa and IIIb15. All the WRKY TFs contain a highly conserved 60 amino acid sequence, the WRKY domain, required for binding target promoters. NAC TF genes also respond to both abiotic and biotic stresses and some are also responsive to ROS16.
Downstream of the TF networks, a plethora of genes activate responses to environmental and developmental signals, although relatively few TF targets have been verified experimentally. Late embryogenesis abundant (LEA) proteins form a family of 51 members divided into nine groups, many of which accumulate in response to abiotic stresses including osmotic stress, cold, drought, salinity and freezing17. Many have been demonstrated to be functionally important in stress responses as their over- and/or ectopic expression results in stress protection18. They are small (10–30 kD) hydrophilic proteins and were first discovered as accumulating in and during seed desiccation, although they are also expressed in vegetative and floral organs19. Although their precise role in plant cells remains largely undiscovered, evidence suggests that they are involved in regulating stress tolerance to cellular dehydration, through the stabilisation of cell membranes, prevention of protein aggregation, nucleic acid homeostasis and redox balancing20,21,22.
Senescence Associated Gene 21 (SAG21/AtLEA5//LEA38; At4g02380) is a member of the Group 3 LEA proteins23. It is mitochondrially located24 in root cells, although it has also been detected in chloroplasts of Arabidopsis leaf protoplasts25, unlike its paralogue (LEA2; At1g02820) which is located in the cytosol. Three gene models have been predicted for the SAG21 protein (TAIR; Supplementary Fig. 1A) and have been identified as transcripts (TAIR26). Unfortunately, proteomic analysis (http://www.peptideatlas.org/) does not extend far enough towards the N terminal to be able to discriminate between the three models, therefore it is not clear whether all three are transcribed in all tissues where the gene is expressed and or translated. SAG21/LEA5 was identified in a complementation screen of the Δ yap1 oxidant sensitive yeast mutant27, suggesting that a key role for SAG21 may be in protection against oxidative stress and reactive oxygen species. Very recently a function for SAG21 in regulating translation in both mitochondria and chloroplasts has been reported which may be a key mechanism for its role in protection against stress25.
The SAG21 gene was originally identified as being up-regulated during leaf senescence, peaking in abundance prior to full senescence then declining with or shortly after the onset of visible senescence28. However, it is also highly expressed in pollen, petals, and roots24,27. As well as developmental regulation, SAG21 expression is up-regulated in response to a variety of abiotic and biotic stresses including cold29, dehydration30, salinity, and wounding24, phloem feeders31, Botrytis cinerea infection24 and both fungal and bacterial elicitors32 as well as being light regulated24,27. SAG21 was also identified in a genome wide association study as associated with adaptation to environmental stress33. Moreover, SAG21 expression is up-regulated by several stress-related plant growth regulators including ethylene, abscisic acid (ABA), jasmonic acid (JA) and salicylic acid (SA) as well as ROS24,27 which may be mediating all or part of the response to stresses. However, although induction of SAG21 in response to dehydration is dependent upon ABA synthesis it is independent of the protein phosphatase ABI127, a key mediator in ABA response pathways. Many LEA proteins can be induced by combinations of several different stresses and stress-related hormones. For example, ABR (ABA-response protein) which belongs to the LEA_4 family is induced by ABA, NaCl, mannitol and darkness34 and the bZIP transcription factor ABI5 binds directly to G-boxes in its promoter.
Given the complexity of the SAG21 responses to the environment and through development, an analysis of its promoter was undertaken using a yeast-1-hybrid (Y1H) screening approach to identify potential TF regulators. Their functional role was then validated by their co-expression in protoplasts with a SAG21p-GUS reporter and analysis of SAG21 expression in mutants. Results show that a wide range of TFs can bind to the SAG21 promoter, and identifies specific members of the ERF, WRKY and NAC TF gene families that are functionally important for SAG21 response to environmental signals.
Results
Gene model 1 for SAG21 seems prevalent in Arabidopsis seedlings
For analysis of the SAG21 promoter region it was first important to define the correct translational start point. The three gene models for AT4G02380 (SAG21/LEA5) predict proteins of three different lengths, one of which (model 2) includes an alternative splice site (Supplementary Fig. 1A). The longest, model 3, includes 359 bp of coding sequence between the intron and the ATG start codon while models 1 and 2 include 74 and 13 bp respectively (Supplementary Fig. 1A) resulting in predicted proteins of 192, 97 and 78 amino acids respectively. To test whether mRNAs consistent with the longest two models were expressed in seedlings, primers were designed to span the ATG start codon in models 1 and 3 and a downstream sequence within the open reading frame. They were tested both on cDNA and genomic DNA. Amplification of a 514 bp PCR product with genomic DNA as template but absence of the expected 414 bp product from cDNA template confirms that, at least in seedlings, both grown under optimal conditions, and stressed, mRNA consistent with model 3 is absent (Supplementary Fig. 1B,C). However, primers spanning the start codon of model 1 and including the intron produced the expected 216 bp product with genomic DNA and the smaller 116 bp product with the cDNA confirming that an mRNA consistent with model 1 is present. An alignment of the model 3 predicted protein with orthologous genes confirms that in other species homology extends up to start of the model 1 predicted sequence, supporting model 1 as likely translated product. Model 1 was therefore used for analysis of the promoter region.
Co-expression of SAG21 with transcription factors, binding of TFs to the SAG21 promoter revealed by ChIP and predicted distribution of cis-elements on the SAG21 promoter
A first analysis of the TFs that might regulate SAG21 was performed using co-expression analysis in PlantPAN3.0. This revealed a high Pearson correlation coefficient (> 0.8) for five TFs linked to development, each from a different family, and 35 TFs from 17 different families linked to stress responses (Supplementary Table 1). Most highly represented TF families associated with stress were Dof (5 TFs) followed by NAC (4), WRKY, MADS and GATA (3). ChIP experiments were also analysed in PlantPAN3.0 to identify TFs known to bind to the 1685 bp upstream of the SAG21 translational start site based on model 1 above. This analysis identified 52 TFs across 32 ChIP experiments. The highest number (21) were identified in an ABA response experiment, 18 TFs in 17 different experiments with seedlings, and eight related to flowering (Supplementary Table 2).
To gain a better overview of TFs that might bind to specific regions of the SAG21 promoter, the 1685 bp upstream of the SAG21 translational start site (based on model 1) were divided into seven overlapping fragments (Fig. 1A). Using PlantPAN3.0, each of the seven overlapping fragments were assessed for the presence of known cis-elements that potentially bind TF families (Table 1), considering the unique portions of each fragment and the overlapping portions separately. Fragment 7 and Fragment 3 contained cis-elements for the largest number of TF families (27 and 22 respectively) while Dof, GATA and ZF-HD families were present on most different portions of the promoter. Cis-elements for 15 different TF families were only found on a single promoter portion.
Analysis of the SAG21 promoter for NAC and WRKY TF binding. (A) Position and sequence of W-boxes (numbered 1–11, position in brackets, in purple) and ANAC TF binding sites (below in green) on the 1685 bp upstream of the translational start of SAG21 based on the PlantPAN 2.0 database, Lindemose et al.35 and Wu et al.36; uppercase letters indicate canonical sequence. Below, the seven fragments used for a yeast-1-hybrid screen of the WRKY transcription factor library. Positively identified NAC (B) and WRKY (C) transcription factors interacting with the SAG21 promoter fragments based on yeast-1-hybrid interactions observed as growth on SD-LTH medium. Promoter fragments resulting in autoactivation of the reporter gene are shaded in grey.
Similar TF families were identified from the three types of analyses but there was remarkably little overlap in the specific TFs identified through co-expression analyses and from ChiP datasets. An experimental approach was therefore employed to complement the bioinformatic analysis.
A yeast-1 hybrid screen of 1500 plant transcription factors with SAG21 gene promoter sequences identified 16 transcription factors
A screen of over 1500 pooled plant TFs using the seven overlapping fragments of the SAG21 promoter (Fig. 1A), spanning 1685 bp upstream of the gene model 1 ATG resulted in the identification of 58 different TFs from 22 families. The ERF family represented the largest group (11 members were identified) followed by HDZIP and TCP (9 members of each) (Supplementary Table 3). Fragment 5 of the SAG21 promoter (− 893 to − 1221) bound most TFs (32) followed by Fragment 1 (-1 to -333) which bound 14, while no TFs bound to Fragment 7, although for this fragment there was substantial autoactivation by the promoter fragment in the absence of the TFs which may have obscured genuine binding.
To confirm the TF-SAG21 promoter interactions following the library screen, 21 individual TFs were paired with the SAG21 promoter fragments. TFs were selected on the basis of their identification in the library screen and availability of the TF in the Y1H vector. Of these, 16 were confirmed to bind to the proximal six SAG21 promoter fragments (Fragments 1 to 6; Table 2, Supplementary Fig. 2). Many of the TFs bound to more or different fragments than the one that they bound to in the initial screen (Supplementary Table 3). Eight of the TFs tested only bound to one fragment, two bound to two fragments and four TFs bound to three promoter fragments. Of the 16 TFs binding to the SAG21 promoter, five were from the ERF family, five were homeodomain leucine zipper TFs, two were TCP TFs and the remainder were one each of bZIP, zinc finger, ARID and MYB families. All the TF families identified and confirmed as Y1H interactors were represented in the predicted cis-elements except the TCP family.
Four NAC TFs bind to the SAG21 promoter by Y1H
The SAG21 promoter contains at least 11 NAC TF predicted binding sites (Fig. 1A) based on the canonical CGT[G/A] sequence and other SAG21 promoter sequences were identified as potential binding sequences of other NAC TFs: including TT(A/C/G)CTT35 and TGCC[GT]36, although more cryptic sites may be present. Two NAC TFs (ANAC102 and ANAC038), were identified in the initial screen of 1500 TFs (which included 62 NAC TFs). Both bound to Fragment 5 (Supplementary Table 3), but neither were confirmed as binding to this fragment of the SAG21 promoter when re-tested. A separate Y1H screen of 94 Arabidopsis NAC TFs only was therefore performed and identified three NAC TFs that bound only to Fragment 1 of the SAG21 promoter: ANAC042, ANAC071 and ANAC038/39, while ANAC013 bound to four different fragments, including Fragment 1 (Fig. 1B).
To investigate further the binding of NAC TFs to Fragment 1, the promoter sequence was analysed for the canonical NAC TF recognition sequence CGT[G/A]. This was not present, but a partial match to the RRYGCCGT sequence which is the core ANAC042 binding sequence36 was located at − 117 bp upstream from the ATG in the SAG21 promoter. This GCCGT sequence was mutated (Fig. 2A) and binding of ANAC042, ANAC071, ANAC038/39, and ANAC013 was re-tested (Fig. 2B). However, the mutation did not appear to affect binding, indicating that the binding site for these NAC TFs is likely to be different and as yet unidentified.
Effect of mutating SAG21 promoter cis elements identified as biding sites for (A,B) NAC and (C,D) WRKY TFs. Regions highlighted in blue represent the W-box sequence, with bases in red below indicating the mutated bases. (B,D) Interactions observed on SD-LTH medium from a Y1H assay of (B) NAC TFs binding to fragment 1 and (D) WRKY TFs binding to fragment 2 and negative controls, using fragment 2 containing mutated W-box 5, mutated W-box 4 and mutated W-box 4 and 5. WRKY TF number is indicated for each interaction image.
Thirteen WRKY transcription factors bind to the SAG21 promoter
A bioinformatic analysis of the 1685 bp upstream of the functional SAG21 ATG start codon revealed eleven W boxes, four of which conformed to the canonical (T)TGAC(C/T) sequence (Fig. 1A). Apart from a non-canonical CGTTG at position − 120 from the ATG which is predicted to bind just one WRKY transcription factor (WRKY1) all the others are predicted to bind between 60 and 72 WRKY transcription factors. As no WRKY TFs were found in the 1500 TF screen (although 61 WRKY TFs were included), a second Y1H library containing 68 WRKY TFs was screened separately, using the same seven SAG21 promoter fragments (Fig. 1A). Based on this Y1H assay, 13 WRKY transcription factors bound to the SAG21 promoter fragments. This is not totally unexpected as the yeast clones were pooled in the 1500 library and therefore not all may have been isolated from the screen. All 13 WRKY TFs bound to Fragment 2 (− 179 to − 479; Fig. 1C), while WRKY63 and WRKY67 also bound to Fragment 1 (− 1 to − 133), Fragment 5 (− 886 to − 1205) and Fragment 6 (− 1126 to − 1445). No WRKY TF were found in this library to bind to SAG21 promoter Fragments 3, 4 or 7.
To further identify whether the canonical W-boxes identified in Fragment 2 (Fig. 2C) are binding sites for the WRKY TFs, mutations were introduced into the sequences of W-box 4 (at − 324 from the ATG) and W-box 5 (at − 362 from the ATG) the SAG21 promoter (Fig. 2C). Mutation of W-box 5 resulted in loss of binding to all of the 13 WRKY TFs except WRKY63 and WRKY67. In contrast, all the 13 previously identified WRKY TFs binding to Fragment 2 interacted with the promoter fragment mutated at W-box 4, except WRKY33 where binding was abolished. WRKY 40, WRKY70 and WRKY52 were included as negative controls as they bound very weakly to the wild type (WT) Fragment 2. Some binding of these TFs is seen to mutated W-box 4 but much weaker than the other WRKY TFs. These results suggest that WRKY33 does not require W-box 4 to bind to the SAG21 promoter while WRKY63 and WRKY67 do not require W-box 5. The double knockout of W-box 4 and W-box 5 abolished all WRKY-TF interactions, except WRKY63 and WRKY67 (Fig. 2D) suggesting that WRKY63 and WRKY67 may bind to an as yet unidentified cryptic site on Fragment 2 of the SAG21 promoter.
Co-expression of SAG21::GUS and pJIT60::WRKY63 in protoplasts increases SAG21 driven expression
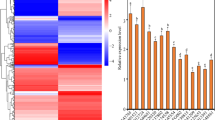
To test whether the binding of WRKY TFs to the SAG21 promoter might be functional in plant cells, Arabidopsis leaf mesophyll protoplasts were co-transformed with a construct in which 1685 bp of the SAG21 promoter was used to drive expression of GUS, and constructs in which the open reading frames of four WRKY TFs were expressed from the 35S promoter. Four WRKY TFs were selected based on their binding to the SAG21 promoter fragments in the Y1H screen and their published stress-responsiveness. WRKY15 expression is up-regulated by reactive oxygen species37, WRKY33 appeared to bind specifically to W box 4 on Fragment 2 of the SAG21 promoter and is involved in abiotic and biotic stress responses38,39, WRKY63 and WRKY67 bound to several fragments of the SAG21 promoter and are involved in responses to drought40 and salt41 respectively. Co-expression of the WRKY63 open reading frame with the SAG21 promoter GUS reporter construct resulted in a significant (P < 0.05) more than twofold increase in expression in the protoplasts compared to the GALDB control (Fig. 3). In contrast, co-expression of SAG21p:GUS with WRKY33, WRKY15 or WRKY67 had no significant effect on SAG21 driven GUS expression.
Functional analysis of WRKY-transcription factor interactions. Relative expression levels derived from fluorescence of SAG21 promoter GUS constructs in response to interactions with transcription factor protein expressing constructs. Both plasmids expressed in Arabidopsis mesophyll protoplasts. The pGPTV-KAN::SAG21(1685)::GUS reporter was assayed with pJIT60::WRKY15, pJIT60::WRKY63 and pJIT60::WRKY67 effectors. Relative expression is normalised to LUC and compared to a GALDB control, with the expression value for GALDB arbitrarily set to 100. n = 3, + SE, *indicates significant difference to GALDB (P < 0.05) based on a student’s T-test.
Bioinformatic prediction of upstream regulators of SAG21, under stress conditions
hCSI (hierarchical Causal Structure Inference) modelling was used to test whether the TFs identified in the three Y1H screens could be part of potential regulatory networks upstream of SAG21 when Arabidopsis plants are under four different stress conditions, based on published microarray datasets of their gene expression. Stresses were: inoculation with Botrytis cinerea, developmental senescence, inoculation with avirulent hrpA or virulent DC3000 strains of Pseudomonas syringae and drought. This modelling resulted in the identification of ABF4, and ANAC039 as potential regulators of SAG21 (probability > 0.4) during both Botrytis and Pseudomonas syringae infection. ANAC042 was identified as a potential regulator during senescence, whereas ANAC071 may regulate SAG21 expression both under drought stress and Botrytis infection (Fig. 4).
hCSI modelling of SAG21 and its upstream potential regulators. Heat map shows the probability (maximum possible value = 1) of each TF regulating SAG21 in a particular stress condition: inoculation with Botrytis cinerea, developmental long day (LD) senescence, inoculation with avirulent hrpA or virulent DC3000 strains of Pseudomonas syringae and drought. Details of treatments in “Methods” section.
SAG21 expression is altered in mutants of ERF, NAC and WRKY transcription factors
To test whether Y1H binding was indicative of a functional role for the TFs in regulating SAG21 expression in planta, expression of SAG21 was analysed in Arabidopsis mutants of five of the TFs identified in the Y1H screens. Plants were grown under optimal conditions or were exposed to stresses or stress-related plant growth regulators already known to affect SAG21 expression24. Expression of SAG21 in ERF1 mutants over-expressor lines was assessed with and without ethylene as this TF is known to mediate responses to ethylene42. Under optimal conditions, SAG21 expression was higher in the erf1 mutant seedlings than in WT Arabidopsis, but SAG21 expression was not affected by ERF1 over-expression (Fig. 5A). Expression of SAG21 in WT seedlings was up-regulated by exposure to ethylene, however in the erf1 mutant background this increase was abolished, and SAG21 expression was in fact down-regulated in response to ethylene compared to untreated controls (Fig. 5A). In the ERF1 over-expressor (OEX) background seedlings, ethylene treatment again down regulated SAG21 expression.
Relative expression of SAG21 in WT and mutants grown under optimal conditions compared to stress or hormone treated (in brackets for each line): (A) erf1-1 seedlings (treated with 100 ppm ethylene), (B) nac042/JUB1 and NAC042OEX seedlings (H2O2 stress for 6 h before harvest), (C) anac071 stems cut and left on the plant for 0, 1 and 3 days, (D) wrky63 seedlings treated with OG (for 1 h before harvest), (E) wrky63 seedlings (ambient dehydration stress for 30 min after harvest), (F) wrky15 amiRNA seedlings (H2O2 stress for 6 h before harvest); mean relative expression ± S.E.; n = 3; different letters indicate significant differences P < 0.05).
Both ANAC042 (JUB136) and WRKY1537 respond to H2O2 and could be mediating the response of SAG21 to ROS. ANAC042 was also identified as a potential regulator of SAG21 during senescence (Fig. 4), which in turn might be mediated by ROS. Exposure of WT seedlings to H2O2 confirmed elevated SAG21 expression levels (Fig. 5B,C24) although the change was not always statistically significant. In seedlings where ANAC042 expression was reduced, this increase was greatly enhanced, although due to variability the change was again not significant (P < 0.05). In amiRNA wrky15 seedlings where WRKY15 expression is reduced, the increase in SAG21 expression elicited by H2O2 treatment was slightly (though not significantly) reduced (Fig. 5B). In agreement with the protoplast co-expression experiments, under optimal growth conditions down-regulation of WRKY15 did not greatly affect SAG21 expression (Fig. 5B). Likewise, down-regulation of ANAC042 in the anac042 (jub1-1) plants did not affect expression of SAG21 under non-stressed conditions (Fig. 5C). However, over-expression of ANAC042 from the 35S promoter appeared to slightly reduce the expression of SAG21 under optimal conditions, and slightly dampened upregulation (although again not significantly, probably due to the variability in expression) (Fig. 5C).
Although ANAC071 was identified as a potential regulator of SAG21 under drought and pathogen stress (Fig. 4), its most important role recognised to date has been in stem wound responses43,44. Expression of SAG21 increased progressively over time when bolting stems were excised, and stored at ambient temperature, hence wounded, as well as exposed to ambient dehydration (Fig. 5D). Although an increase in SAG21 expression over time was also seen in anac071 excised stems after one day of storage, it was abolished after 3 days.
WRKY63 (also known as ABO3) is important in drought and ABA signalling40, and also responds to salicylic acid (SA)15. Since SAG21 expression is also upregulated by SA and pathogens24 as well as oligogalacturonide (OG) elicitors32, the effect of OG elicitors on expression of SAG21 was tested in wrky63 insertion mutant seedlings under non-stressed and ambient dehydration stress treatment and in response to OG treatment. Dehydration stress slightly increased SAG21 expression in seedlings, although not significantly (Fig. 5E). Under optimal conditions, expression of SAG21 in wrky63 mutants was elevated compared to WT (although with substantial variability across replicates), in agreement with the co-expression in protoplasts. A more dramatic effect of WRKY63 on SAG21 expression was seen when wrky63 mutant seedlings were challenged with OG elicitor (Fig. 5F). In the WT there was a dramatic upregulation of SAG21 expression which was significantly reduced, though not abolished in wrky63 mutant seedlings.
Discussion
The large number of TFs predicted to bind to, and identified as binding to, the SAG21 promoter indicates a complex regulatory network upstream of this gene. However, there was not complete agreement between TF families predicted to bind, likely TF regulators from co-expression analyses and those discovered by Y1H or found in ChIP databases. This suggests that cryptic TF binding sites are present which are not identified by the tools available such as PlantPan3.0, and, of course, the wide Y1H screen using pools of TFs may easily have missed interactors. However, overall, 33 potential regulators of SAG21 expression were identified here using the Y1H approach and importantly, confirmed by repeating the Y1H. These included TFs from nine families: 13 WRKY TFs, five ERF and five HB TFs, four NACs, two TCP and one each of ABF, ZAT, HMG-box and MYB. Of these, four WRKYs were further tested in protoplast co-expression assays. Only WRKY63 was able to activate SAG21:GUS reporter expression. This may reflect the limitation of this approach as only leaf mesophyll cells are represented, and other TFs required for activation of SAG21 promoter activity may not be represented. The functional role of five TFs (one ERF, two WRKY and two NAC family) were tested based on the availability of mutant lines and provide a complex picture of SAG21 promoter activation.
Many ERF transcription factors are involved in the response to abiotic stresses9 and may be mediators of stress signals to SAG21. Here we showed that five ERF TFs were able to bind to regions of the SAG21 promoter in a Y1H assay and may be involved in regulating the response of SAG21 to pathogens. ERF5 is a positive regulator of JA mediated defence and a negative regulator of SA signalling45 and ERF14 is also involved in pathogen responses46. ERF10 is a repressor up-regulated in response to SA + JA47 so that may implicate it in pathogen responses, although it was also upregulated during leaf senescence48. ERF15 is a positive regulator of ABA responses49 and likely mediates both salinity and drought responses. ERF15 may share a binding site with ERF1 as it also bound the − 1 to − 333 promoter fragment, while the other three ERFs bound the promoter further upstream. The lack of consistency between ERFs co-expressed with SAG21, identified through the Y1H screen, and identified though ChIP studies as potential direct regulators of SAG21 likely reflects the complexity of SAG21 regulation given that ERF1 regulation may only be important under specific stress conditions or developmental stages. Other ERFs identified in the ChIP studies (AP2 in inflorescences, ERF115 in roots and BBM in seedlings) may require plant factors to bind to the SAG21 promoter or may have been missed in the wide TF screen. It has been suggested that some TFs are not always processed correctly in yeast50 possibly resulting in false-negatives during the Y1H assay, causing potentially significant TF interactions to be missed.
ERF1 was selected for further study as it plays several roles in both stress responses and development. For example, in plant pathogen interactions ERF1 integrates JA and ET signalling42 as it responds synergistically to the two hormones. ERF1 also responds to salinity, heat and drought stresses30. In development it mediates ET-induced repression of root growth51 and has a role in regulating flowering time52. SAG21 was upregulated in 35S::ERF1 plants under drought stress30 and indeed the SAG21 promoter contains a single GCC element (GCCGCC) in inverse orientation 70 bp upstream from the ATG start codon. This is included in Fragment 1 which was one of the two SAG21 promoter fragments that bound ERF1 in the Y1H screen. The finding that SAG21 expression increases in WT Arabidopsis when challenged with ethylene is thus consistent. However, the upregulation of SAG21 expression in erf1 mutants in the absence of ethylene suggests that ERF1 may also be acting as a repressor of SAG21 expression when ethylene is not present. ERF1 was previously reported as a repressor of FLOWERINGLOCUS T (FT) to delay floral initiation51, so a repression of SAG21 is not inconsistent with its known functions. The lack of upregulation in ERF1-OEX seedlings under non-stressed conditions is also consistent with the data from Cheng et al.30. The higher expression of SAG21 in erf1 mutants may reflect other signalling pathways or regulatory components, but the lack of upregulation of SAG21 in the erf1 mutant suggests that the upregulation of SAG21 by ethylene may be mediated primarily through ERF1.
The two TCP TFs confirmed as binding to the SAG21 promoter (TCP4 and TCP8) may regulate its expression both during development and in response to stress. TCP4 regulates leaf development53 and represses petal greening54 although it is also involved in the repression of leaf area under high temperature55 so has a role in mediating abiotic stress signals. TCP8 is involved in SA signalling as part of host immunity56 and also in modulating brassinosteroid responses57. Although the SAG21 promoter was not identified in a ChIP experiment with TCP8, SAG21 was down-regulated in tcp8 mutants57, showing that there is an effect of TCP8 on SAG21 expression. The Y1H data here suggest that the interaction could be direct and might have been missed in the ChIP screen.
Of the five homeodomain leucine zipper TFs identified in the initial Y1H screen (Supplementary Table 3) and confirmed as Y1H interactors, HB5 was identified in a ChIP experiment with ABA treated seedlings and indeed HB5 is a likely ABA signalling regulator in seedlings58. HB12 is involved in root and leaf development under non-stressed conditions, and seed production under drought conditions59. HB13 is involved in seed to seedling development60 but also in drought and salinity tolerance61. HB23 is involved in co-regulating blue light signalling and growth62. HB52 is also involved in light responses, and their integration with nitrogen status63, but is also ethylene-responsive and controls primary root elongation64. These transcription factors may therefore be important in regulating SAG21 during plant growth while responding to environmental signals and are worthy of further investigation.
Of the other four TFs identified in the initial 1500 TF Y1H screen, and confirmed as Y1H interactors, ABF4 was also identified as a potential upstream regulator of SAG21 in the hCSI modelling during pathogen attack. ABF4 is involved in ABA signalling65 and ABA was discovered to be a negative regulator of B. cinerea infection66. Of the remaining three TFs confirmed as Y1H interactors with the SAG21 promoter from the initial screen, ZAT10/STZ is induced by a wide range of abiotic stresses including salt, cold, ABA and dehydration67 and was also recently identified as a hub gene in plant pathogen interactions68 and MYB67 is highly upregulated during seed dormancy69. These TFs may therefore mediate some of the upregulation of SAG21 expression elicited by ABA27. Finally, AT1G76110 is part of the light response network70 and therefore may be involved in the known light regulation of SAG2124,27.
NAC TFs play important roles in mediating stress responses8. Two NAC TFs were identified from the ChIP seq database data as binding to the SAG21 promoter: ANAC032 and ANAC102 in ABA challenged seedlings (Supplementary Table 271) while six other NAC TFs were co-expressed with SAG21 (Supplementary Table 1): these include ANAC033, ANAC70 and ANAC100 under environmental stress conditions and ANAC81, ANAC87 and ANAC91 under hormone treatments8. Of these only ANAC102 was not included in the Y1H screen. Four different NAC TFs from those co-expressed or detected in ChIP experiments (ANAC013, ANAC038/39, ANAC042 and ANAC071) were confirmed to bind to the SAG21 promoter in the − 1 to − 133 region (Fragment 1) by Y1H. However, no canonical motifs for NAC TFs, i.e., CGT[G/A]35 were found in this region of the SAG21 promoter. This binding may be explained because the CGT[G/A] canonical sequence72 is seemingly not universal across all NAC TFs. For example, ANAC042 which binds to the SAG21 promoter, bound to a different core sequence RRYGCCGT36. Since mutation of the GCCGT sequence in the SAG21 promoter did not abolish binding of any of the four NAC TFs, there must be other cryptic sites in this sequence that enable binding of these NAC TFs. ANAC038/39 was identified in the hCSI modelling as a potential regulator of SAG21 expression during Botrytis and Pseudomonas infection, and bound to Fragment 1 of the SAG21 promoter. Expression of this NAC TF is upregulated by Botrytis infection73 as is SAG2124 so this role would warrant further verification.
The functional role of two of the NAC TFs identified in the SAG21 promoter Y1H screen was assessed. ANAC042, also known as JUB1, is a negative regulator of leaf senescence and is strongly upregulated by H2O236. The increased upregulation of SAG21 expression in the anac042 mutant background when treated with H2O2 suggests that ANAC042 may be acting as a negative regulator of SAG21 expression by ROS. This is consistent with the role of ANAC042 as a repressor in the regulation of anthocyanin biosynthetic genes36. ANAC042 is also important as a negative regulator of senescence36 and our modelling suggested that this TF might be a regulator of SAG21 during this stage of development. SAG21 is only very transiently expressed during developmental senescence24,28. Nevertheless, over-expression of SAG21 delayed leaf senescence, so it may play a role in mitigating the effects of rising ROS during senescence.
A much clearer effect on SAG21 expression was detected in anac071 mutants where wound-induced SAG21 upregulation was significantly impaired (Fig. 5D). ANAC071 is involved in wounding repair: its expression is activated 1–3 days after wounding in Arabidopsis stems and anac071 mutants show impaired vascular tissue regeneration following wounding43. The function of SAG21 in wound repair is unclear, but it is strongly and rapidly (within minutes) upregulated by wounding also in leaves24. This long-term upregulation of SAG21expression 3 days after wounding suggests that SAG21 may have a role both in early and later wounding responses. Wounding elicits the production of ethylene within 20 min of wounding74 and ANAC071 upregulation around wounding sites is thought to be regulated by both auxin accumulation and ethylene. As SAG21 expression is also ethylene induced, this might be part of the signalling pathway (Fig. 6). However, SAG21 expression following wounding may also be induced by OGs via an increase in H2O2. Wounding elicits a rise in H2O2 activated by OG release from the damaged cells75. H2O2 accumulates gradually at the site of wounding76 acting as a signalling molecule to activate defence responses, and hence could activate SAG21 expression, since SAG21 expression is induced by ROS24,27.
Model of upstream regulators of SAG21 expression based on the Y1H screens and real time qPCR. Biotic and abiotic stress signals activate plant hormones and signalling molecules which in turn activate TFs belonging to several families. These TFs activate and repress SAG21 expression depending on the condition.
As well as NACs, WRKY TFs are another major plant TF family involved in the regulation of both biotic and abiotic stress signals6,8. Eight WRKY TF genes were co-expressed with SAG21, including WRKY15 that was co-expressed under environmental and biotic stress as well as hormone treatment (Supplementary Table 3). Only two of these TFs, WRKY15 and WRKY33, bound to the SAG21 promoter in the Y1H screen. Although activation of SAG21 expression by either WRKY15 or WRKY33 was not confirmed in protoplast co-expression assays, induction of SAG21 expression by H2O2 was reduced (though not significantly) in the wrky15 mutant indicating that WRKY15 may be contributing to the upregulation of SAG21 expression by H2O2. As well as these two WRKY TFs, a further eleven were identified as binding to the SAG21 promoter through Y1H analysis. All 13 WRKY TFs identified in the screen bound to Fragment 2 (Fig. 1) but only two of these bound to Fragment 1. As there is a large overlap of 155 bp between these two fragments, this suggests that the region from − 333 to − 479 bp upstream of the ATG is playing a major role in WRKY TF recognition. Indeed, all these WRKY TFs appeared to bind to W-box 5 (located at − 362) but not W-box 4 (located at − 324) apart from WRKY33 that binds to both cis-elements. In addition, WRKY63 and WRKY67 also bind to some other cryptic site(s) within Fragment 2 of the SAG21 promoter and were the only two WRKY TFs that bound to more than one promoter fragment, interacting with a total of four separate fragments of the 1685 bp SAG21 promoter. Different WRKY TFs have differing specificities for variants of the W-box11 and binding of WRKY TFs to anomalous sites has also been reported77,78. For example, WRKY70 can bind to the YGACTTTT sequence77 and this sequence is also present at positions − 38 and − 368 in the SAG21 promoter, found in Fragment 1 and Fragment 2, respectively. However, regardless of the presence of this motif in both fragments, WRKY70 was not identified in our Y1H screens. WRKY63 and WRKY67 are phylogenetically similar and are members of the IIIa sub-group15 that share a highly conserved amino acid region outside of the WRKY domain, hence may also share a yet undiscovered alternative binding sequence within target promoters.
Co-expression studies of the SAG21p-GUS reporter construct and 35S-WRKY candidate genes in protoplasts confirmed the activation of SAG21 expression by WRKY63. WRKY63 also known as ABO3 is important in drought and ABA signalling40. However, SAG21 expression was higher under both non-stress and dehydration stress treatments in the wrky63 mutant background (although with increased variability) suggesting that it might actually be acting as a repressor. WRKY63 can act as a repressor of MYB28/29 under water deficit79 hence this role is plausible. WRKY63 mutation had a stronger effect on the response of SAG21 to the OG elicitor. Upregulation of SAG21 expression by OG has been previously reported32 and is consistent with its upregulation by both fungal and bacterial pathogens24 independently of a hypersensitive response. SAG21 expression is also upregulated in response to flg22 a peptide elicitor derived from bacterial flagellin32,80. WRKY63 expression responds to SA and is amongst the IIIa sub-group WRKY genes that require SA accumulation to induce their expression following infection with Pernospora parasitica15. Surprisingly, the study by Denoux et al.32 did not show that WRKY63 expression responded to OGs. The partial dependence of SAG21 upregulation on WRKY63 in response to the OG elicitor may therefore be mediated by SA induction of WRKY63. Of note is that WRKY63 is involved in the activation of BCS1, a mitochondrial protein, in response to flg22 treatment14 and the expression pattern of WRKY63 indicated a role as a positive regulator of mitochondrial proteins upon stress. Since SAG21 is a mitochondrial protein that also responds to ROS, the role of WRKY63 as an activator is consistent. The contrasting results on the role of WRKY63 on SAG21 expression likely reflect different roles of this TF under different conditions.
The lack of SAG21 promoter activation in protoplasts when co-expressed with WRKY15, WRKY33 and WRKY67 could indicate that the Y1H binding to the SAG21 promoter is not functionally significant or that activation only occurs under specific stresses or developmental stages. A role of WRKY15 in regulating the response of SAG21 to ROS would be consistent with its role in mediating communication between mitochondrial stress responses37, as well as WRKY33, a key regulator of pathogen responses38, and WRKY67 which is activated by salt stress41. Other WRKY TFs binding to the SAG21 promoter are also worthy of further investigation through the analysis of SAG21 expression in mutant lines.
In conclusion, three TFs belonging to three different families have been identified as regulators of SAG21 expression, ERF1, ANAC071 and WRKY63. Their functional role has been confirmed through analysis of Arabidopsis TF mutant lines. A further two TFs: WRKY15 and ANAC042 may also regulate SAG21 expression, although further verification is needed. A tentative model of their interaction is proposed (Fig. 6). However, further work will be needed to assess the functional significance of other TF interactions with SAG21 identified here through Y1H screens. Analysing SAG21 expression under different developmental stages with and without different stresses and/or stress combinations, in combination with ChIP analysis of candidate TFs and a detailed analysis of the SAG21 promoter is essential to identify crucial promoter elements involved in its transcriptional regulation. This will enable the regulatory network controlling this complex promoter to be disentangled from the bottom up and top down.
Methods
Analysis of transcription factor binding sites and cloning of promoter fragments
PlantPAN3.081 was used to identify TFs that co-express with SAG21/AtLEA5/LEA38 (At4g02380) and ChIP experiments showing binding of TFs to 1685 bp upstream of the SAG21 ATG start codon.
PlantPAN3.081 was also used to find predicted cis-elements in this 1685 bp region. The 1685 bp was divided into seven overlapping fragments of approximately 300 bp based on this analysis for use in Y1H screens. Fragments were amplified using gene-specific primers (Supplementary Table 4) and cloned into the pHISLEU2 vector (Invitrogen) using restriction enzymes EcoRI and MluI. All cloned fragments were sequenced (deposited to Genbank with accession codes OR513440–OR513446) and then transformed into the Y187 (MATα) yeast strain (Clontech) to form baits for the Y1H screens.
Y1H screens
Three libraries of TFs were screened containing full-length TF open reading frames fused to the N-terminal GAL4 activation domain in the vector pDEST22 (Invitrogen). The first library comprised approximately 1500 TFs (REGIA + REGULATORS; RR Library82 and was kindly donated by the authors. The TF library was transformed into the yeast strain AH109 (MATa) and pooled as in Ref.50. The other two libraries comprised 94 NAC TFs and 75 WRKY TFs, and consisted of sequence verified constructs (Supplementary Table 550).
The libraries were screened essentially as in Ref.50. Yeast clones were grown on SD-Trp (library) and SD-Leu (bait) and mated on YPDA medium, then replica plated onto SD-Leu-Trp, SD-Leu-Trp-His with the addition of 1–100 mM 3-amino-1,2,4-triazol (3AT). Successful mating was checked by growth on SD-Leu-Trp. After overnight incubation and replica cleaning, plates were incubated for 4 days. Positive colonies (growing on SD-Leu-Trp-His medium) were patched onto selective plates. Multiple yeast clones of each promoter fragment were tested against the libraries to ensure consistent results. To identify the interacting TF from the 1500 TF library, yeast colonies were treated with 20 mM NaOH at 100 °C for 10 min, and then subjected to nested colony PCR. The first amplification was with primers SABR447 and SABR448; for the second amplification, PCR product was diluted 10× and amplified with primers SABR150 and SABR4506 (primer sequences in Supplementary Table 4). PCR products from the second PCR were identified by partial sequencing following purification with a QIAquick kit (Qiagen). TFs were identified using WuBLAST within TAIR. Screening of NAC and WRKY TF libraries was carried out in the same way except sequencing of positive colonies was not required.
To verify the Y1H positive TF interactions, plasmids carrying individual TFs were transformed into the Y187 yeast strain and mated with the bait-carrying yeast strain as above. Three independent yeast transformants for each promoter fragment bait construct were tested.
Mutation of cis-elements in promoter fragments
W-box sequences and the NAC binding sequence (TGCC[GT]36) were mutated using a QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). Primers are listed in Supplementary Table 4. Mutated clones were sequenced (deposited to Genbank with accession codes OR513447–OR513450), and a second round of mutagenesis was applied to introduce mutations into both W-box sequences within the same promoter fragment. For each Y1H assay, four separate promoter clones (containing the mutation) were used, along with a non-mutated promoter fragment clone as a positive control.
Analysis of transcription factor binding in protoplasts
Protoplasts were isolated from Arabidopsis leaves and transformed transiently essentially as in Ref.83. Protoplast integrity and number were checked using a light microscope. Protoplasts were co-transformed With both pGPTV-Kan-SAG21(1685)::GUS (as described in Ref.24) and control plasmid pJIT60-35S::LUC or TF plasmids pJIT60-35S::WRKY15, pJIT60-35S::WRKY63, or pJIT60-35S::WRKY67. All the pJIT60-35S clones were created from pDONRZeo entry clones via Gateway cloning (Invitrogen). Clones were sequenced to verify integrity of the TF coding sequences. A luciferase assay kit (Promega) was used to detect LUC and a methyl-umbelliferone (MUG) based β-glucuronidase assay for detection of GUS, and both reporters were analysed with a TECAN M2000 pro Fluorimeter. pJIT60-35S::GAL4DB was used as a negative control instead of the TF clone. LUC luminescence was used to verify transformation, GUS fluorescence was used to assess promoter activation. Assays were repeated three times. Data were normalised against LUC and then presented as relative expression to the GALDB control.
hCSI modelling
Hierarchical Causal Structure Inference (hCSI) analysis84 was based on the detailed time course microarray data sets generated under the PRESTA project following leaf senescence over 3 weeks85, infection with Botrytis cinerea over 48 h73, Pseudomonas syringae infection over 17.5 h86 and 6 h of high light treatment87.
Plant material and growth
All Arabidopsis thaliana genotypes were in the Columbia background. A homozygous erf1 knock out (KO) line was selected from segregating lines containing a T-DNA insertion (GK-850A03/N481507) obtained from NASC (Nottingham Arabidopsis Stock Centre). ERF1 over-expressor (OEX) 35S::ERF1-1 (AT3G23240/N6143) was also obtained from NASC. A homozygous artificial miRNA (amiRNA) line for WRKY15 (AT2G23320) that knocks down WRKY15 expression was as described in Ref.37. T-DNA insertion lines for wrky63 (At1g66600; SALK_068280C) were obtained from NASC. Insertion position in the knockouts were verified by PCR (Supplementary Figs. 3–6), insertion knocks out expression of WRKY6340. Anac071 (SALK_012841C) seeds were as in Ref.43. A T-DNA insertion line of anac042 (also known as jub1-1; AT2G43000; SALK_036474) in which expression was knocked down and of 35S::ANAC042 over-expression line were as in Ref.36.
All seeds were surface sterilised and stratified in sterilised distilled water for 24 h at 4 °C. For growth on Petri dishes seeds were sown onto sterile 1× Murashige and Skoog (MS) basal salt mix, 1% agar, 1% sucrose, pH 5.5–5.7 and sealed with MicroporeTM tape. Seedlings were germinated and grown in an environmentally controlled growth chamber at 21 °C under long days (16 h day, 8 h night). For obtaining stem tissue, seeds were sown onto freeze sterilized John Innes soil mix (soil: sand at 3:1 ratio) and grown in a growth chamber (16 h light, 21 °C) until bolting. Following bolting, plants were grown for an additional 7 days, and stem tissue was then excised.
Stress treatments
Seedlings were pre-treated with 12 h light (90–100 μmol photons m−2 s−2) before treatment to repress expression of the SAG21 gene which is light regulated24,27. Ethylene (100 ppm) was injected into sealed Petri dishes containing 7 d old seedlings and then left for 24 h in the light. As a dehydration stress treatment 14 d old seedlings were removed from Petri dishes, placed on Whatmann filter paper for 30 min and exposed to air flow in a laminar air flow cabinet. For the control treatment, Petri dishes were left in the growth chamber for 30 min88. Hydrogen peroxide stress treatment of 14 d old seedlings was carried out by submerging seedlings in 1× MS liquid medium with 10 mM H2O2 for 6 h as previously described36, control seedlings were left on the plates for the 6 h period. To analyse wound response in stem tissue, stems of soil grown flowering plants were excised just above the rosette leaves, placed into sterile 15 mL falcon tubes and left for 1 or 3 days at room temperature. Stem tissue was then excised from just below the first cauline leaves. To test elicitor responses, seedlings were challenged with a galacturonan oligosaccharide mixture (OG, DP10/DP15elicitor, Elicityl). Whole rosettes of 4-week-old plants grown in soil were vacuum infiltrated in a 200 µM solution of OG for 5 min. After 1 h of treatment, leaves were then snap-frozen in liquid nitrogen. Control leaves were totally immersed in water. Whole seedlings, leaves or stem sections from three biological replicates of the control and stress treatments were harvested and frozen in liquid nitrogen and stored at − 80 °C.
Analysis of gene expression by real time qPCR
Plant material (100–150 mg) was ground into a fine powder with liquid nitrogen and RNA isolation was carried out using an RNeasy Plant Mini Kit (QIAGEN) or using TRI-Reagent (Sigma). Residual genomic DNA was removed using RQ1 DNase (Promega). First strand cDNA was synthesized from RNA samples using M-MLV RNase H Reverse Transcriptase (Promega) using oligo dT (Promega). Real time qPCR used 60 ng of cDNA, 0.4 μL of SAG21 forward and reverse primers (10 μM) (Supplementary Table 4), 10 μL of 2× qPCRBIOSyGreen Mix Lo-ROX (PCR Biosystems), in a 20 μL reaction volume, and was conducted using a Light Cycler 96 (Roche) machine. PCR thermal profiling conditions were: 95 °C for 120 s, 95 °C for 30 s, 60 °C for 30 s, 72.0 °C for 30 s for 35 cycles followed by melting curve analysis from 60 to 98 °C to check for primer specificity and primer dimers. Gene expression analysis was carried out using the relative comparative method89 using the 2−ΔΔct method.
Plant materials
Use of plants in the present study complies with international, national and institutional guidelines. No seeds were obtained as collections from wild populations of Arabidopsis.
Data availability
All key data are provided in the Supplementary Files. Sequences have been deposited with Genbank (Accession Numbers OR513440–OR513450).
References
Zhang, H., Zhu, J., Gong, Z. & Zhu, J. K. Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119 (2022).
Hilker, M. & Schmülling, T. Stress priming, memory, and signalling in plants. Plant Cell Environ. 42, 753–761 (2019).
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E. & Mittler, R. Abiotic and biotic stress combinations. New Phytol. 203, 32–43 (2014).
Anwar, K., Joshi, R., Dhankher, O. P., Singla-Pareek, S. L. & Pareek, A. Elucidating the response of crop plants towards individual, combined and sequentially occurring abiotic stresses. Int. J. Mol. Sci. 22, 6119 (2021).
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J. P. & Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113 (2019).
Jiang, J. et al. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101 (2017).
Abdullah-Zawawi, M. R. et al. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice. Sci. Rep. 11, 19678 (2021).
Nuruzzaman, M., Sharoni, A. M. & Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248 (2013).
Xie, Z., Nolan, T. M., Jiang, H. & Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10, 228 (2019).
Wang, Z., Niu, Y. & Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 22, 6125 (2021).
Ciolkowski, I., Wanke, D., Birkenbihl, R. P. & Somssich, I. E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92 (2008).
He, L., Xu, J., Wang, Y. & Yang, K. Transcription factor ANAC074 binds to NRS1, NRS2, or MybSt1 element in addition to the NACRS to regulate gene expression. Int. J. Mol. Sci. 19, 3271 (2018).
Li, W., Pang, S., Lu, Z. & Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 9, 1515 (2020).
Van Aken, O., Zhang, B., Law, S., Narsai, R. & Whelan, J. AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol. 162, 254–271 (2013).
Kalde, M., Barth, M., Somssich, I. E. & Lippok, B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signalling pathways. Mol. Plant Microbe Interact. 16, 295–305 (2003).
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K. & Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103 (2012).
Hundertmark, M. & Hincha, D. K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 9, 118 (2008).
Hernández-Sánchez, I. E. et al. LEAfing through literature: Late embryogenesis abundant proteins coming of age—Achievements and perspectives. J. Exp. Bot. 73, 6525–6546 (2022).
Bies-Etheve, N. et al. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 67, 107–124 (2008).
Bremer, A., Wolff, M., Thalhammer, A. & Hincha, D. K. Folding of intrinsically disordered plant LEA proteins is driven by glycerol-induced crowding and the presence of membranes. FEBS J. 284, 919–936 (2017).
Wallmann, A. & Kesten, C. Common functions of disordered proteins across evolutionary distant organisms. Int. J. Mol. Sci. 21, 2105 (2020).
Graether, S. P. Proteins involved in plant dehydration protection: The late embryogenesis abundant family. Biomolecules 12, 1380 (2022).
Avelange-Macherel, M.-H., Candat, A., Neveu, M. & Tolleter, D. Decoding the divergent subcellular location of two highly similar paralogous LEA proteins. Int. J. Mol. Sci. 19, 1620 (2018).
Salleh, F. M. et al. A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ. 35, 418–429 (2012).
Karpinska, B. et al. Late embryogenesis abundant (LEA)5 regulates translation in mitochondria and chloroplasts to enhance growth and stress tolerance. Front. Plant Sci. 13, 875799 (2022).
Zhang, R. et al. A high-resolution single-molecule sequencing-based Arabidopsis transcriptome using novel methods of Iso-seq analysis. Genome Biol. 23, 149 (2022).
Mowla, S. B. et al. Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant (LEA)-like protein in oxidative stress tolerance. Plant J. 48, 743–756 (2006).
Weaver, L. M., Gan, S., Quirino, B. & Amasino, R. M. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37, 455–469 (1998).
Vyse, K., Schaarschmidt, S., Erban, A., Kopka, J. & Zuther, E. Specific CBF transcription factors and cold-responsive genes fine-tune the early triggering response after acquisition of cold priming and memory. Physiol. Plant 174, e13740 (2022).
Cheng, M. C., Liao, P. M., Kuo, W. W. & Lin, T. P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582 (2013).
Kempema, L. A., Cui, X., Holzer, F. M. & Walling, L. L. Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 143, 849–865 (2007).
Denoux, C. et al. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1, 423–445 (2008).
Fournier-Level, A. et al. A map of local adaptation in Arabidopsis thaliana. Science 334, 86–89 (2011).
Su, M. et al. The LEA protein, ABR, is regulated by ABI5 and involved in dark-induced leaf senescence in Arabidopsis thaliana. Plant Sci. 247, 93–103 (2016).
Lindemose, S. et al. A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res. 42, 7681–7693 (2014).
Wu, A. et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24, 482–506 (2012).
Vanderauwera, S. et al. AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 20113–20118 (2012).
Zheng, Z., Qamar, S. A., Chen, Z. & Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605 (2006).
Jiang, Y. & Deyholos, M. K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105 (2009).
Ren, X. et al. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429 (2010).
Ma, S., Gongand, Q. & Bohnert, H. J. Dissecting salt stress pathways. J. Exp. Bot. 57, 1097–1107 (2006).
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J. & Solano, R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178 (2003).
Asahina, M. et al. Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 16128–16132 (2011).
Matsuoka, K. et al. Wound-inducible ANAC071 and ANAC096 transcription factors promote cambial cell formation in incised Arabidopsis flowering stems. Commun. Biol. 4, 369 (2021).
Moffat, C. S. et al. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7, e35995 (2012).
Huang, P. Y., Catinot, J. & Zimmerli, L. Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 67, 1231–1241 (2016).
Caarls, L. et al. Assessing the role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes. Plant Cell Physiol. 58, 266–278 (2017).
Balzadeh, S., Riano-Pachón, D. M. & Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 10, 63–75 (2008).
Lee, S., Bee, S., Lee, S. J. & Kim, S. Y. AtERF15 is a positive regulator of ABA response. Plant Cell Rep. 34, 71–81 (2015).
Hickman, R. et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 75, 26–39 (2013).
Mao, J. L. et al. Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet. 12, e1005760 (2016).
Chen, Y., Zhang, L., Zhang, H., Chen, L. & Yu, D. ERF1 delays flowering through direct inhibition of FLOWERING LOCUS T expression in Arabidopsis. J. Integr. Plant Biol. 63, 1712–1723 (2021).
Challa, K. R., Rath, M. & Nath, U. The CIN-TCP transcription factors promote commitment to differentiation in Arabidopsis leaf pavement cells via both auxin-dependent and independent pathways. PLoS Genet. 15, e1007988 (2019).
Zheng, X. et al. Arabidopsis transcription factor TCP4 represses chlorophyll biosynthesis to prevent petal greening. Plant Commun. 3, 100309 (2022).
Saini, K., Dwivedi, A. & Ranjan, A. High temperature restricts cell division and leaf size by coordination of PIF4 and TCP4 transcription factors. Plant Physiol. 190, 2380–2397 (2022).
Wang, X. et al. TCP transcription factors are critical for the coordinated regulation of ISOCHORISMATE SYNTHASE 1 expression in Arabidopsis thaliana. Plant J. 82, 151–162 (2015).
Spears, B. J. et al. Class I TCP transcription factor AtTCP8 modulates key brassinosteroid-responsive gene. Plant Physiol. 190, 1457–1473 (2022).
Johannesson, H., Wang, Y., Hanson, J. & Engström, P. The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Mol. Biol. 51, 719–729 (2003).
Re, D. A., Capella, M., Bonaventure, G. & Chan, R. L. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 14, 150 (2014).
Silva, A. T., Ligterink, W., Ribone, P. A., Chan, R. L. & Hilhorst, H. W. M. A predictive co-expression network identifies novel genes controlling the seed-to-seedling phase transition in Arabidopsis thaliana. Plant Physiol. 170, 2218–2231 (2016).
Cabello, J. V. & Chan, R. L. The homologous homeodomain-leucine zipper transcription factors HaHB1 and AtHB13 confer tolerance to drought and salinity stresses via the induction of proteins that stabilize membranes. Plant Biotech. J. 10, 815–825 (2012).
Perrella, G. et al. ZINC-FINGER interactions mediate transcriptional regulation of hypocotyl growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E4503–E4511 (2018).
Ariga, T., Sakuraba, Y., Zhuo, M., Yang, M. & Yanagisawa, S. The Arabidopsis NLP7-HB52/54-VAR2 pathway modulates energy utilization in diverse light and nitrogen conditions. Curr. Biol. 32, 1–10 (2022).
Miao, Z.-Q. et al. Arabidopsis HB52 mediates the crosstalk between ethylene and auxin by transcriptionally modulating PIN2, WAG1, and WAG2 during primary root elongation. Plant Cell 30, 2761–2778 (2018).
Kang, J. Y., Choi, H. I., Im, M. Y. & Kim, S. Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357 (2002).
Liu, S., Kracher, B., Ziegler, J., Birkenbihl, R. P. & Somssich, I. E. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife 4, e07295 (2015).
Sakamoto, H. et al. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136, 2734–2746 (2004).
Biniaz, Y. et al. Transcriptome meta-analysis identifies candidate hub genes and pathways of pathogen stress responses in Arabidopsis thaliana. Biology 11, 1155 (2022).
Barrero, J. M. et al. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 61, 611–622 (2010).
Bobrovskikh, A. V., Zubairova, U. S., Bondar, E. I., Lavrekha, V. V. & Doroshkov, A. V. Transcriptomic data meta-analysis sheds light on high light response in Arabidopsis thaliana L.. Int. J. Mol. Sci. 23, 4455 (2022).
Song, L. et al. A transcription factor hierarchy defines an environmental stress response network. Science 354, 1550 (2016).
Puranik, S., Sahu, P. P., Srivastava, P. S. & Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381 (2012).
Windram, O. et al. Arabidopsis defense against Botrytis cinerea: Chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24, 3530–3557 (2012).
Lin, Z., Zhong, S. & Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 60, 3311–3336 (2009).
Galletti, R., Ferrari, S. & De Lorenzo, G. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157, 804–814 (2011).
Prasad, A., Sedlarova, M., Balukova, A., Rac, M. & Pospisil, P. Reactive oxygen species as a response to wounding: In vivo imaging in Arabidopsis thaliana. Front. Plant Sci. 10, 1660 (2020).
Machens, F., Becker, M., Umrath, F. & Hehl, R. Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol. Biol. 84, 371–385 (2013).
Van Verk, M. C., Bol, J. F. & Linthorst, H. J. M. WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 11, 89 (2011).
Salehin, M. et al. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 10, 4021 (2019).
Zipfel, C. et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767 (2004).
Chow, C.-N. et al. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 47, D1155–D1163 (2019).
Castrillo, G. et al. Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS ONE 6, e21524 (2011).
Yoo, S. D., Cho, Y.-H. & Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Penfold, C. A., Buchanan-Wollaston, V., Denby, K. J. & Wild, D. L. Nonparametric Bayesian inference for perturbed and orthologous gene regulatory networks. Bioinformatics 28, i223–i241 (2012).
Breeze, E. et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894 (2011).
Lewis, L. A. et al. Transcriptional dynamics driving MAMP-triggered immunity and pathogen effector-mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv. tomato DC3000. Plant Cell 27, 3038–3064 (2015).
Alvarez-Fernandez, R. et al. Time-series transcriptomics reveals a BBX32-directed control of acclimation to high light in mature Arabidopsis leaves. Plant J. 107, 1363–1386 (2021).
Kilian, J. et al. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363 (2007).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
The authors thank Prof Mueller-Roeber and Prof Balazadeh for providing us with seed for the nac042 mutant and the NAC042 over-expressor line. They also thank Prof Van Breusegem for providing us with seed of amiRNAwrky15.
Funding
The funding was provided by Cardiff University, Malaysian government (PhD studentship), IAESTE UK (UK/12/200/05f), ERASMUS (Placement Project), BBSRC (BBSRC BB/F005806/1), Biotechnology and Biological Sciences Research Council and Higher Education Commission, Pakistan (IRSIP 49 BMS 01). KVE is now an employee of AstraZeneca, and may or may not own stock options.
Author information
Authors and Affiliations
Contributions
KVE, ER, SN, BW, FMS, IG, LE, CT, AH, SB, RJH, KP all contributed to the experimental work and data analysis. CH, BdG, KD, VB-W and HJR were closely involved in experimental design and data analysis. HJR drafted the manuscript and all authors approved it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Evans, K.V., Ransom, E., Nayakoti, S. et al. Expression of the Arabidopsis redox-related LEA protein, SAG21 is regulated by ERF, NAC and WRKY transcription factors. Sci Rep 14, 7756 (2024). https://doi.org/10.1038/s41598-024-58161-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-58161-0
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.