Featured
-
-
Article
| Open Access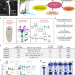 An evolutionarily-conserved Wnt3/β-catenin/Sp5 feedback loop restricts head organizer activity in Hydra
An evolutionarily-conserved Wnt3/β-catenin/Sp5 feedback loop restricts head organizer activity in Hydra
Hydra regenerate various body parts on amputation by activation of the appropriate organiser, but how head formation is controlled is unclear. Here, the authors identify the transcription factor Sp5 as restricting head formation, by being activated by beta-catenin and then acting as a repressor of Wnt3.
- Matthias C. Vogg
- , Leonardo Beccari
- & Brigitte Galliot
-
Article
| Open Access Human genome-wide measurement of drug-responsive regulatory activity
Human genome-wide measurement of drug-responsive regulatory activity
Quantification of genomic responses to environmental stimuli by current genome-scale assays is limited to indirect measurements or requires knowledge of the transcription factors involved. Here, the authors use genome-wide high-throughput reporter assays to agnostically map enhancer activity in response to glucocorticoid treatment across the human genome.
- Graham D. Johnson
- , Alejandro Barrera
- & Timothy E. Reddy
-
Article
| Open Access Molecular structure of promoter-bound yeast TFIID
Molecular structure of promoter-bound yeast TFIID
Transcription preinitiation complex assembly begins with the recognition of the gene promoter by the TATA-box Binding Protein-containing TFIID complex. Here the authors present a Cryo-EM structure of promoter-bound yeast TFIID complex, providing a detailed view of its subunit organization and promoter DNA contacts.
- Olga Kolesnikova
- , Adam Ben-Shem
- & Gabor Papai
-
Article
| Open Access The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family
The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family
AUXIN RESPONSE FACTORS (ARFs) are a family of plant-specific transcriptional factors involved in auxin signaling. Here, the authors adapt DAP-seq technology to show the binding landscape of 14 maize ARFs and reveal class-specific binding properties and transcriptional coordination by ARFs from different classes.
- Mary Galli
- , Arjun Khakhar
- & Andrea Gallavotti
-
Article
| Open Access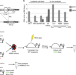 EVI1 overexpression reprograms hematopoiesis via upregulation of Spi1 transcription
EVI1 overexpression reprograms hematopoiesis via upregulation of Spi1 transcription
Chr3q26 rearrangements cause overexpression of EVI1 and associate with myeloid neoplasms, but the mechanism behind this association is unclear. Here, using a novel mouse model they show that EVI1 causes premalignant myeloid expansion with suppression of other lineages through upregulation of Spi1/PU.1.
- Edward Ayoub
- , Michael P. Wilson
- & Archibald S. Perkins
-
Article
| Open Access Divergent wiring of repressive and active chromatin interactions between mouse embryonic and trophoblast lineages
Divergent wiring of repressive and active chromatin interactions between mouse embryonic and trophoblast lineages
The role of the genome structure in the establishment of the embryonic and trophoblast lineages is still not well understood. Here the authors perform promoter capture Hi-C in mouse trophoblast and embryonic stem cells and find divergent networks of repressive and active chromatin interactions between the two lineages.
- Stefan Schoenfelder
- , Borbala Mifsud
- & Miguel R. Branco
-
Article
| Open Access Repurposing of promoters and enhancers during mammalian evolution
Repurposing of promoters and enhancers during mammalian evolution
Enhancers and promoters are different types of regulatory elements with shared architectural and functional features. Here the authors perform integrated cross-mammalian analyses of DNase hypersensitivity, chromatin modification and transcriptional data, to provide evidence of regulatory repurposing during evolution.
- Francesco N. Carelli
- , Angélica Liechti
- & Henrik Kaessmann
-
Article
| Open Access Analysis of chromatin accessibility uncovers TEAD1 as a regulator of migration in human glioblastoma
Analysis of chromatin accessibility uncovers TEAD1 as a regulator of migration in human glioblastoma
The intrinsic drivers of glioblastoma (GBM) migration are still poorly understood. Here the authors purify GBM stem cells (GSCs) from patients and profile chromatin accessibility in these cells, identifying TEAD1 as a regulator of migration in human glioblastoma.
- Jessica Tome-Garcia
- , Parsa Erfani
- & Nadejda M. Tsankova
-
Article
| Open Access Jungle Express is a versatile repressor system for tight transcriptional control
Jungle Express is a versatile repressor system for tight transcriptional control
Tightly regulated promoters with strong inducibility and scalability are highly desirable for biological applications. Here the authors describe ‘Jungle Express’, a EilR repressor-based broad host system activated by cationic dyes.
- Thomas L. Ruegg
- , Jose H. Pereira
- & Michael P. Thelen
-
Article
| Open Access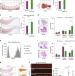 A metabolic interplay coordinated by HLX regulates myeloid differentiation and AML through partly overlapping pathways
A metabolic interplay coordinated by HLX regulates myeloid differentiation and AML through partly overlapping pathways
HLX transcription factor regulates haematopoietic stem and progenitor cell (HSPC) differentiation and is overexpressed in acute myeloid leukemia. Here the authors show that HLX overexpression leads to myeloid differentiation block in zebrafish and human HSPCs by direct regulation of metabolic pathways.
- Indre Piragyte
- , Thomas Clapes
- & Eirini Trompouki
-
Article
| Open Access Human pluripotent reprogramming with CRISPR activators
Human pluripotent reprogramming with CRISPR activators
CRISPRa is an attractive tool for cellular reprogramming due to its multiplexing capacity and direct targeting of genomic loci. Here the authors demonstrate the reprogramming of human fibroblasts into iPSCs, which is enhanced by targeting a conserved Alu-motif.
- Jere Weltner
- , Diego Balboa
- & Timo Otonkoski
-
Article
| Open Access Postnatal DNA demethylation and its role in tissue maturation
Postnatal DNA demethylation and its role in tissue maturation
Here the authors show that a large fraction of the tissue-specific methylation pattern is generated postnatally. These changes, which occur in response to hormone signaling, appear to play a major role in the regulation of gene expression and tissue maturation in the liver.
- Yitzhak Reizel
- , Ofra Sabag
- & Howard Cedar
-
Article
| Open Access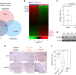 Estrogen-related receptor gamma functions as a tumor suppressor in gastric cancer
Estrogen-related receptor gamma functions as a tumor suppressor in gastric cancer
Very little is known regarding the molecular mechanisms involved in gastric cancer development. Here the authors show estrogen-related receptor gamma (ESRRG) is a tumor suppressor in gastric cancer and suggest the mechanism of this tumor suppression function involves the inhibition of Wnt signaling.
- Myoung-Hee Kang
- , Hyunji Choi
- & Yun-Yong Park
-
Article
| Open Access A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing
A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing
Previous studies suggest that DNA methylation is the main mechanism to silence endogenous retroviruses (ERVs) in somatic cells. Here the authors provide evidence that distinctive sets of ERVs are silenced by Setdb1 in different types of somatic cells, suggesting a general function in ERV silencing.
- Masaki Kato
- , Keiko Takemoto
- & Yoichi Shinkai
-
Article
| Open Access Bcl11b is essential for licensing Th2 differentiation during helminth infection and allergic asthma
Bcl11b is essential for licensing Th2 differentiation during helminth infection and allergic asthma
Th2 cells are critical in the resolution of helminth infection and allergy. Here the authors show that Bcl11b is required to license the Th2 program of helper T cell differentiation and restrict alternate lineage gene expression using in vivo models of helminth infection and asthma.
- Kyle J. Lorentsen
- , Jonathan J. Cho
- & Dorina Avram
-
Article
| Open Access Characterization of the enhancer and promoter landscape of inflammatory bowel disease from human colon biopsies
Characterization of the enhancer and promoter landscape of inflammatory bowel disease from human colon biopsies
Many SNPs associated with inflammatory bowel disease are located in non-coding genomic regions. Here, the authors perform CAGE-sequencing on descending colon biopsies of Crohn’s disease and ulcerative colitis patients to map transcription start sites and enhancer activity for analysis of regulatory regions.
- Mette Boyd
- , Malte Thodberg
- & Albin Sandelin
-
Article
| Open Access HOXA9 inhibits HIF-1α-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development
HOXA9 inhibits HIF-1α-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development
Hypoxia-inducible transcription factor HIF-1α promotes glycolysis allowing cell survival under stress. Here the authors show, using both cell lines and animal models, that in cutaneous squamous cell carcinoma HOXA9 acts as a tumor suppressor and inhibits glycolysis by associating with CRIP2 to repress HIF-1α binding to target genes.
- Liang Zhou
- , Yinghui Wang
- & Zhenhua Ding
-
Article
| Open Access Cryptic glucocorticoid receptor-binding sites pervade genomic NF-κB response elements
Cryptic glucocorticoid receptor-binding sites pervade genomic NF-κB response elements
Glucocorticoid receptor (GR) regulates immunity and inflammation but the mechanisms by which GR represses proinflammatory genes are still being debated. Here the authors use a multidisciplinary approach and show that GR binds to a cryptic site within genome-wide NFκB DNA response elements to repress pro-inflammatory genes.
- William H. Hudson
- , Ian Mitchelle S. de Vera
- & Eric A. Ortlund
-
Article
| Open Access GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network
GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network
In breast cancer GATA3 is known to be frequently mutated, but the function of these mutations is unclear. Here, the authors utilise CRISPR-Cas9 to model frame-shift mutations in zinc finger 2 of GATA3, highlighting that GATA3 mutation can have gain- or loss-of function effects in breast cancer.
- Motoki Takaku
- , Sara A. Grimm
- & Paul A. Wade
-
Article
| Open Access MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state
MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state
Breast cancer tumors originating from mammary luminal epithelial cells are highly heterogeneous. Here, the authors show MYC-driven tumor initiation is reliant on cell reprogramming via an epigenetic program which leads to mammary luminal epithelial cells acquiring basal/stem cell-like properties.
- Vittoria Poli
- , Luca Fagnocchi
- & Alessio Zippo
-
Article
| Open Access A regulatory circuit of two lncRNAs and a master regulator directs cell fate in yeast
A regulatory circuit of two lncRNAs and a master regulator directs cell fate in yeast
Transcription of lncRNAs is known to regulate local gene expression. Here the authors describe how orchestrated transcription of two contiguous lncRNAs facilitates a regulatory circuit by which the master regulator for entry into meiosis, IME1, directs the decision to enter meiosis in yeast.
- Fabien Moretto
- , N. Ezgi Wood
- & Folkert J. van Werven
-
Article
| Open Access SOD3 improves the tumor response to chemotherapy by stabilizing endothelial HIF-2α
SOD3 improves the tumor response to chemotherapy by stabilizing endothelial HIF-2α
Tumour vasculature influences drug delivery. Here, the authors show that SOD3 re-expression enhances doxorubicin delivery and effects through normalization of tumour vasculature via the HIF-2a/VE-cadherin pathway.
- Emilia Mira
- , Lorena Carmona-Rodríguez
- & Santos Mañes
-
Article
| Open Access AhR and SHP regulate phosphatidylcholine and S-adenosylmethionine levels in the one-carbon cycle
AhR and SHP regulate phosphatidylcholine and S-adenosylmethionine levels in the one-carbon cycle
Methyl metabolites in the one-carbon cycle, such as phosphatidylcholines and S-adenosylmethionine, play a role in hepatic triglyceride regulation. Here Kim et al. show that AhR and SHP are both involved in the expression of several key enzymes of one-carbon metabolism, with the former regulating them early after feeding and the latter inhibiting AhR at later stages.
- Young-Chae Kim
- , Sunmi Seok
- & Jongsook Kim Kemper
-
Article
| Open Access HoxC5 and miR-615-3p target newly evolved genomic regions to repress hTERT and inhibit tumorigenesis
HoxC5 and miR-615-3p target newly evolved genomic regions to repress hTERT and inhibit tumorigenesis
The expression of telomerase catalytic subunit hTERT is frequently upregulated in many cancers. Here, the authors show HoxC5 and miR-615-3p can negatively regulate hTERT to impede tumorigenesis by targeting the newly evolved cis-regulatory genomic elements of hTERT.
- TingDong Yan
- , Wen Fong Ooi
- & Shang Li
-
Article
| Open Access The transcript cleavage factor paralogue TFS4 is a potent RNA polymerase inhibitor
The transcript cleavage factor paralogue TFS4 is a potent RNA polymerase inhibitor
Transcript cleavage factors such as eukaryotic TFIIS assist the resumption of transcription following RNA pol II backtracking. Here the authors find that one of the Sulfolobus solfataricus TFIIS homolog—TFS4—has evolved into a potent RNA polymerase inhibitor potentially involved in antiviral defense.
- Thomas Fouqueau
- , Fabian Blombach
- & Finn Werner
-
Article
| Open Access Developmental YAPdeltaC determines adult pathology in a model of spinocerebellar ataxia type 1
Developmental YAPdeltaC determines adult pathology in a model of spinocerebellar ataxia type 1
Ataxin-1, linked to spinocerebellar ataxia type 1, is known to interact with the orphan nuclear receptor RORα. Here, Fujita and colleagues show that genetic supplementation of RORα-interacting protein YAPdeltaC during early development can rescue the adult pathologies of SCA1 mouse model.
- Kyota Fujita
- , Ying Mao
- & Hitoshi Okazawa
-
Article
| Open Access Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells
Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells
The repertoire of tissue-specific distal regulators of gene transcription enhancers defines homeostasis or disease. Here, the authors reveal the enhancer and super-enhancer signature of vascular smooth muscle cells under normal and angiotensin II stimuli, providing new insight into the transcriptional regulation of vascular pathologies.
- Sadhan Das
- , Parijat Senapati
- & Rama Natarajan
-
Article
| Open Access Computational design of small transcription activating RNAs for versatile and dynamic gene regulation
Computational design of small transcription activating RNAs for versatile and dynamic gene regulation
The structural basis of RNA-based gene control offers the possibility of de novo design. Here the authors present a computational design approach for Small Transcription Activating RNAs a bacterial RNA regulator that allows for versatile and dynamic control of genes, pathways and genetic circuits.
- James Chappell
- , Alexandra Westbrook
- & Julius B. Lucks
-
Article
| Open Access Integrating evolutionary and regulatory information with a multispecies approach implicates genes and pathways in obsessive-compulsive disorder
Integrating evolutionary and regulatory information with a multispecies approach implicates genes and pathways in obsessive-compulsive disorder
Obsessive-compulsive disorder (OCD) is a neuropsychiatric disorder with symptoms including intrusive thoughts and time-consuming repetitive behaviors. Here Noh and colleagues identify genes enriched for functional variants associated with increased risk of OCD.
- Hyun Ji Noh
- , Ruqi Tang
- & Kerstin Lindblad-Toh
-
Article
| Open Access A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development
A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development
Zbtb16-encoded transcription factor PLZF directs the differentiation of multiple innate and innate-like cell lineages, but how Zbtb16 itself is regulated remains unclear. Here the authors show, using CRISPR gene editing, ATAC-seq and ChIP-seq, that specific Runx1-bound enhancer elements critically modulate lineage-dependent expressions of PLZF.
- Ai-Ping Mao
- , Isabel E. Ishizuka
- & Albert Bendelac
-
Article
| Open Access A long-range cis-regulatory element for class I odorant receptor genes
A long-range cis-regulatory element for class I odorant receptor genes
“Each olfactory sensory neuron expresses a single odorant receptor gene from either class I or class II genes. Here, the authors identify an enhancer for mouse class I genes, that is highly conserved, and regulates most class I genes expression by acting over ~ 3 megabases within the whole cluster.”
- Tetsuo Iwata
- , Yoshihito Niimura
- & Junji Hirota
-
Article
| Open Access Dynamic regulation of canonical TGFβ signalling by endothelial transcription factor ERG protects from liver fibrogenesis
Dynamic regulation of canonical TGFβ signalling by endothelial transcription factor ERG protects from liver fibrogenesis
The transcription factor ERG is key to endothelial lineage specification and vascular homeostasis. Here the authors show that ERG balances TGFβ signalling through the SMAD1 and SMAD3 pathways, protecting the endothelium from endothelial-to-mesenchymal transition and consequent liver fibrosis in mice via a SMAD3-dependent mechanism.
- Neil P. Dufton
- , Claire R. Peghaire
- & Anna M. Randi
-
Article
| Open Access An HDAC3-PROX1 corepressor module acts on HNF4α to control hepatic triglycerides
An HDAC3-PROX1 corepressor module acts on HNF4α to control hepatic triglycerides
HDAC3 is a critical mediator of hepatic lipid metabolism and its loss leads to fatty liver. Here, the authors characterize the liver HDAC3 interactome in vivo, provide evidence that HDAC3 interacts with PROX1, and show that HDAC3 and PROX1 control expression of genes regulating lipid homeostasis.
- Sean M. Armour
- , Jarrett R. Remsberg
- & Mitchell A. Lazar
-
Article
| Open Access HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells
HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells
HIV insertions in hematopoietic cells are enriched in BACH2 or MLK2 genes, but the selective advantages conferred are unknown. Here, the authors show that BACH2 and additionally STAT5B are activated by viral insertions, generating chimeric mRNAs specifically enriched in T regulatory cells favoring their persistence.
- Daniela Cesana
- , Francesca R. Santoni de Sio
- & Eugenio Montini
-
Article
| Open Access Identifying DNase I hypersensitive sites as driver distal regulatory elements in breast cancer
Identifying DNase I hypersensitive sites as driver distal regulatory elements in breast cancer
Cancer driver mutations can occur within noncoding genomic sequences. Here, the authors develop a statistical approach to identify candidate noncoding driver mutations in DNase I hypersensitive sites in breast cancer and experimentally demonstrate they are regulatory elements of known cancer genes.
- Matteo D′Antonio
- , Donate Weghorn
- & Kelly A Frazer
-
Article
| Open Access The endothelial transcription factor ERG mediates Angiopoietin-1-dependent control of Notch signalling and vascular stability
The endothelial transcription factor ERG mediates Angiopoietin-1-dependent control of Notch signalling and vascular stability
Vascular maturation and stability is regulated by Notch and Angiopoietin-1/Tie2 signalling. Here, Shahet al. show that the transcription factor ERG coordinates the Ang1, Notch and Wnt/β-catenin pathways to promote vascular maturation and stability.
- A. V. Shah
- , G. M. Birdsey
- & A. M. Randi
-
Article
| Open Access Zinc-dependent regulation of zinc import and export genes by Zur
Zinc-dependent regulation of zinc import and export genes by Zur
Zinc homeostasis in most bacteria is achieved by a set of regulators, each responding to a certain level of intracellular zinc. Here the authors show that, inStreptomyces coelicolor, the Zur regulator modulates the expression of genes for zinc import and export over a large range of zinc concentrations.
- Seung-Hwan Choi
- , Kang-Lok Lee
- & Jung-Hye Roe
-
Article
| Open Access Endothelial LRP1 regulates metabolic responses by acting as a co-activator of PPARγ
Endothelial LRP1 regulates metabolic responses by acting as a co-activator of PPARγ
LDL receptor-related protein 1 (LRP1) is an endocytic receptor involved in cell signalling and energy homeostasis. Here Maoet al. demonstrate that endothelial Lrp1 modulates lipid and glucose metabolism by binding the nuclear receptor Pparγ and promoting its transcriptional activity.
- Hua Mao
- , Pamela Lockyer
- & Xinchun Pi
-
Article
| Open Access EcR recruits dMi-2 and increases efficiency of dMi-2-mediated remodelling to constrain transcription of hormone-regulated genes
EcR recruits dMi-2 and increases efficiency of dMi-2-mediated remodelling to constrain transcription of hormone-regulated genes
InDrosophila, ecdysone-regulated genes are bound by a heterodimer of ecdysone receptor (EcR) and Ultraspiracle. Here, the authors provide evidence of a non-canonical complex containing EcR and the nucleosome remodeller dMi-2 that limits transcription of ecdysone-regulated genes.
- Judith Kreher
- , Kristina Kovač
- & Alexander Brehm
-
Article
| Open Access The histone H3K9 methyltransferase SUV39H links SIRT1 repression to myocardial infarction
The histone H3K9 methyltransferase SUV39H links SIRT1 repression to myocardial infarction
The molecular pathways regulating the cardioprotective activity of deacetylase sirtuin-1 are unknown. Here, Yanget al. show that histone H3K9 methyltransferase SUV39H and HP1gamma cooperatively methylate H3K9 on the sirtuin-1 promoter and inhibit sirtuin-1 transcription, and show that inhibition of SUV39H in mice is cardioprotective.
- Guang Yang
- , Xinyu Weng
- & Aijun Sun
-
Article
| Open Access Pleckstrin homology domain-containing protein PHLDB3 supports cancer growth via a negative feedback loop involving p53
Pleckstrin homology domain-containing protein PHLDB3 supports cancer growth via a negative feedback loop involving p53
p53 is an oncosuppressor regulating several genes at the transcriptional level. Here, the authors identify a negative feedback loop between PHLDB3 and p53; PHLDB3 is a transcriptional target of p53 which facilitates MDM2-mediated p53 ubiquitination and degradation, impacting on tumorigenesis.
- Tengfei Chao
- , Xiang Zhou
- & Hua Lu
-
Article
| Open Access Multiple myeloma risk variant at 7p15.3 creates an IRF4-binding site and interferes with CDCA7L expression
Multiple myeloma risk variant at 7p15.3 creates an IRF4-binding site and interferes with CDCA7L expression
Genome wide association studies have identified multiple risk loci for multiple myeloma. Here, the authors show that the expression of CDCA7Lis associated with patient survival and expression of the gene is influenced by a risk variant at 7p15.3, which creates a transcription factor binding site for IRF4.
- Ni Li
- , David C. Johnson
- & Richard S. Houlston
-
Article
| Open Access Repression of RNA polymerase by the archaeo-viral regulator ORF145/RIP
Repression of RNA polymerase by the archaeo-viral regulator ORF145/RIP
How archaeal viruses perturb host transcription machinery is poorly understood. Here, the authors provide evidence that the archaeo-viral transcription factor ORF145/RIP targets host RNA polymerase, repressing its activity.
- Carol Sheppard
- , Fabian Blombach
- & Finn Werner
-
Article
| Open Access AKAP95 regulates splicing through scaffolding RNAs and RNA processing factors
AKAP95 regulates splicing through scaffolding RNAs and RNA processing factors
The chromatin-associated protein AKAP95 is known for its chromatin-related functions including enhancing transcription. Here the authors show that AKAP95 interacts with the splicing regulatory factors as well as RNAs to regulate the inclusion of exons and pre-mRNA splicing.
- Jing Hu
- , Alireza Khodadadi-Jamayran
- & Hao Jiang
-
Article
| Open Access Genome-wide compendium and functional assessment of in vivo heart enhancers
Genome-wide compendium and functional assessment of in vivo heart enhancers
Identification of non-coding variants has outstripped our ability to annotate and interpret them. Dickel et al. present a compendium of over 80,000 putative human heart enhancers and demonstrate that two conserved enhancers are required for proper cardiac function in mice.
- Diane E. Dickel
- , Iros Barozzi
- & Len A. Pennacchio
-
Article
| Open Access Capture of associated targets on chromatin links long-distance chromatin looping to transcriptional coordination
Capture of associated targets on chromatin links long-distance chromatin looping to transcriptional coordination
Chromatin architecture is a key regulator of transcriptional processes, however current methods to investigate it have technical limitations. Here, the authors describe a novel chromatin capture technique, CATCH, which can be used to identify and characterize complex genomic interaction networks.
- Ryan J. Bourgo
- , Hari Singhal
- & Geoffrey L. Greene
-
Article
| Open Access Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity
Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity
To modulate gene expression, the glucocorticoid receptor binds to response elements (RE) that vary in sequence. Here, the authors show that RE sequences can modulate glucocorticoid receptor structure and activity, which might provide regulatory specificity towards individual target genes.
- Stefanie Schöne
- , Marcel Jurk
- & Sebastiaan H. Meijsing
-
Article
| Open Access Metal-responsive promoter DNA compaction by the ferric uptake regulator
Metal-responsive promoter DNA compaction by the ferric uptake regulator
The Fur protein regulates transcription of bacterial genes in response to metal ions. Here, the authors show that the Fur protein from Helicobacter pylorirepresses transcription by iron-responsive oligomerization and DNA compaction, encasing the transcriptional start site in a macromolecular complex.
- Davide Roncarati
- , Simone Pelliciari
- & Alberto Danielli
-
Article
| Open Access Golgi membrane fission requires the CtBP1-S/BARS-induced activation of lysophosphatidic acid acyltransferase δ
Golgi membrane fission requires the CtBP1-S/BARS-induced activation of lysophosphatidic acid acyltransferase δ
CtBP1-S/BARS is required for fission of endomembrane compartments including the Golgi. Here the authors show that CtBP1-S/BARS activates a trans-Golgi lysophosphatidic acid acyltransferase that catalyses the production of phosphatidic acid and is required for fission of the post-Golgi carrier membrane.
- Alessandro Pagliuso
- , Carmen Valente
- & Alberto Luini

 Diverse motif ensembles specify non-redundant DNA binding activities of AP-1 family members in macrophages
Diverse motif ensembles specify non-redundant DNA binding activities of AP-1 family members in macrophages