
Protists give coral colonies their colour. Credit: Sirachai Arunrugstichai/Getty
For risky research with great potential, dive deep
Multimillion-dollar experiment generates tools to study marine microorganisms.
25 February 2019
Nature Index 360°
Take a breath, and thank the oceans for it. Then breathe out, and thank the oceans again. This is not the next big meditation fad. It is recognition of the deep blue’s vital role in sustaining life.
Marine organisms capture more than half of the carbon dioxide sequestered daily through photosynthesis, creating an equal exchange of oxygen. Many of these organisms are tiny, single-celled, greatly understudied creatures known as protists.
But the work of marine protists doesn’t end there. Having captured the CO2, protists then become a primary source of carbon in the food chain. Many protists also produce volatile compounds known as dimethyl sulfides, which help form clouds. And one group ensures that coral colonies retain their healthy appearance.
This is only what is known so far about these pervasive floaters. There is, no doubt, much yet to discover about their ecological contribution. But, how?
Human knowledge of basic biology has been gleaned mostly from some hundred organisms that are easy to genetically manipulate. But such bench favourites as the house mouse, the fruit fly and the zebrafish are inadequate for understanding the diverse workings of the millions of species that comprise life on Earth. And they are especially poor analogues for ocean-dwelling protists.
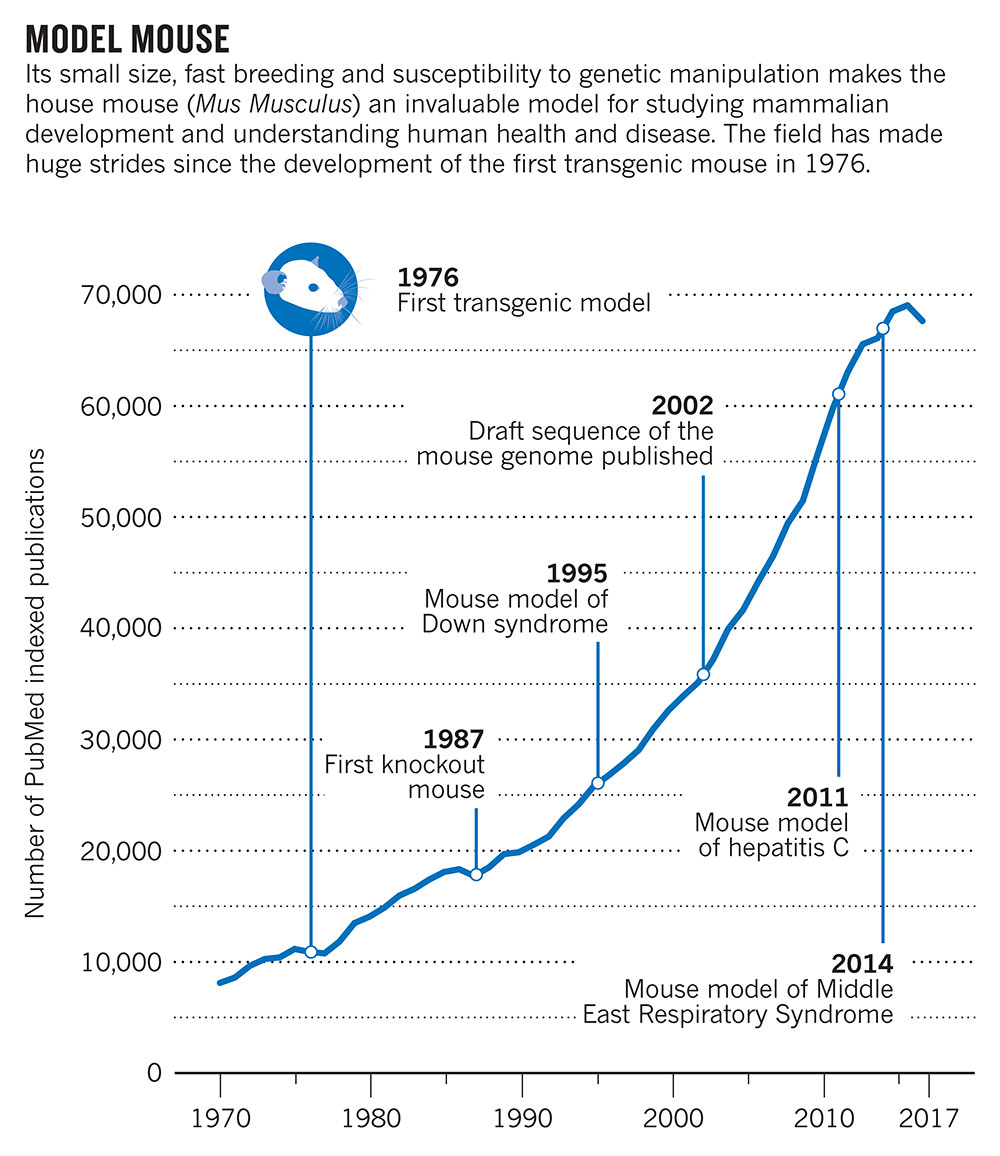
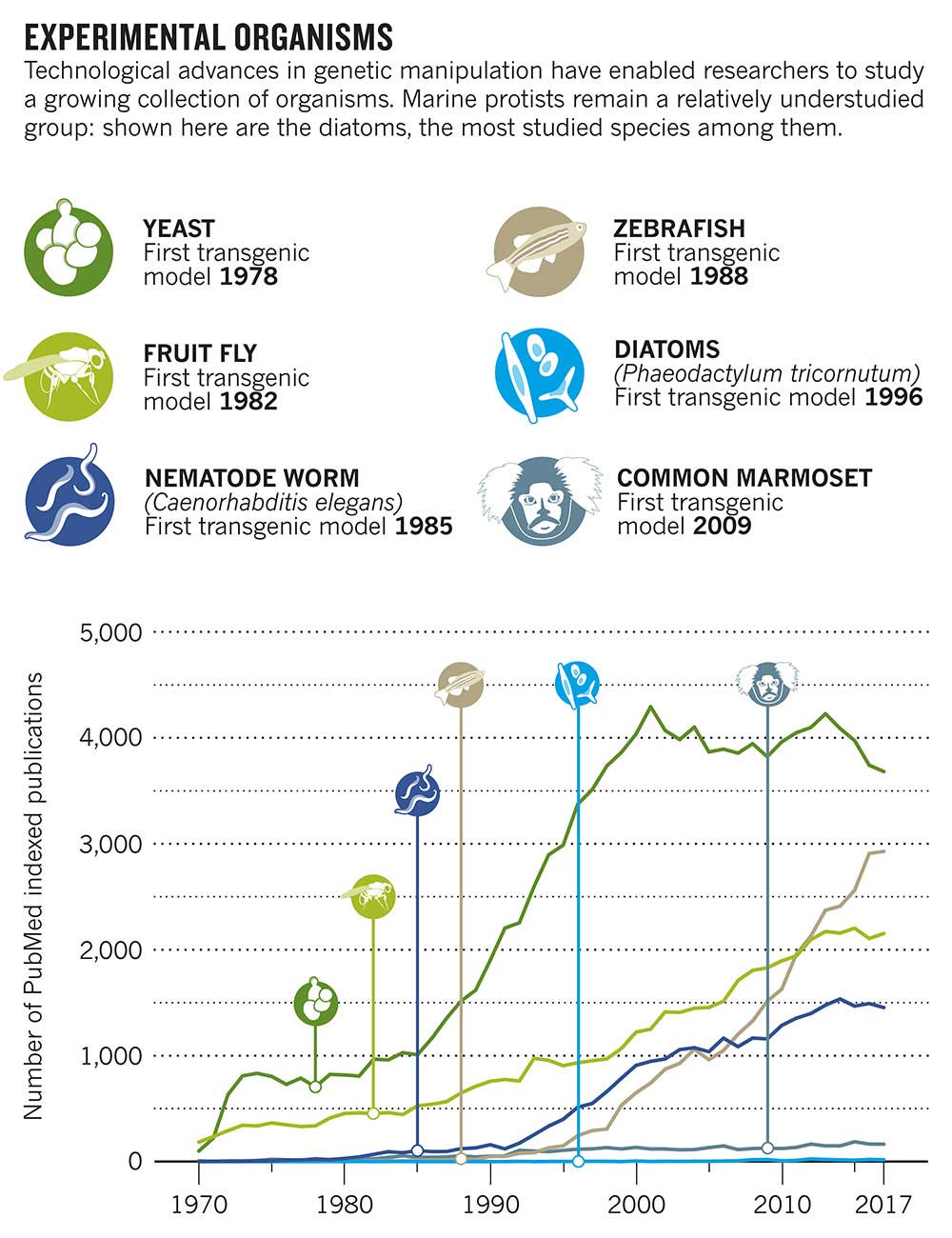
A recent US$15 million funding experiment by the San Francisco-based Gordon and Betty Moore Foundation has ditched these metaphorical hammers, wrenches and saws of research to invest in entirely new instruments for studying marine protists. It has scrapped the mouse, fly and fish for the diatom, diplonemid and dinoflagellate — protist groups found in the oceans.
The initiative seeks to answer myriad questions about these microorganisms. Scientists want to know how marine protists will respond to skyrocketing greenhouse gas emissions, which could have knock-on effects on the climate and fish production. They want to know what causes the devastating bleaching of coral reefs.
Some scientists believe that all of life on Earth is at the mercy of the humble protist. “The happiness of protists in the ocean will to a large extent decide the fate of this planet,” says molecular pathologist, Julius Lukeš, a Moore grant recipient, who heads the Institute of Parasitology, Biology Centre, Czech Academy of Sciences.
The discovery process might uncover a wealth of genetic potential for industrial use, as has been the case with other well-studied organisms. Escherichia coli bacteria are used as low-cost factories for producing therapeutic proteins, and yeast strains are used to ferment beer, bread and sake.
But the marine protist project was a gamble, given that only a handful of their genomes had been successfully tweaked when it started.
“This type of research would be difficult to get funded from national funding agencies because it is simply too risky — it is not easily publishable, there are no visibly strong papers on the subject, and it is not patentable,” says Lukeš. While the research is of great service to the broader scientific community, the personal gains are small, he says: even if, after an entire year’s toil, a poorly studied protist was successfully manipulated, the results would not be of interest to a high-end journal.
“We even considered funding technicians instead of postdocs because, if it didn’t work, they [the postdocs] would have wasted all those years,” says Adam Jones, a programme officer for the Moore Foundation.
In an attempt to mitigate the risk, the funders took a novel approach. They spread the funding across multiple teams, with grants worth a few hundred thousand dollars. They also mandated that grantees openly share their step-by-step recipes for genetic manipulation with each other — the victories with the defeats.
The project has supported more than 100 scientists in 33 institutions around the world, including Israel, Italy, Japan, Chile, the United States, and Spain. It has led to some 18 published articles so far, and created an open and collaborative community.
Saltwater bonanza
The term protist is a symptom of neglect. It was long used as a catchall for whatever couldn’t be categorized in the other kingdoms for eukaryotes, whose cells contain a nucleus. Scientists have come to recognize the deficiency of this label as advances in DNA sequencing and molecular evolution reveal the vast diversity among protists. They are as genetically different from each other as animals are different from plants, says molecular biologist, Adrian Clarke, at the Department of Biological and Environmental Sciences, University of Gothenburg in Sweden, another Moore grant recipient. “They are very anciently related.”
Some of the best known protists are pathogens, including the mould Phytophthora infestans, which caused widespread potato blight during the Great Hunger in Ireland in the mid-19th century, and Plasmodium, responsible for malaria. These terrestrial scourges have been the primary targets of protist research, with little attention paid to those offshore.
“More people study sparrows than all marine protists combined,” says Lukeš, who has recently shifted focus from parasitic protists implicated in diseases such as sleeping sickness and leishmaniasis, to the marine, non-pathological variety. “Nobody cares about the invisible organisms in the ocean — they were there billions of years ago and will be there billions of years from now.”
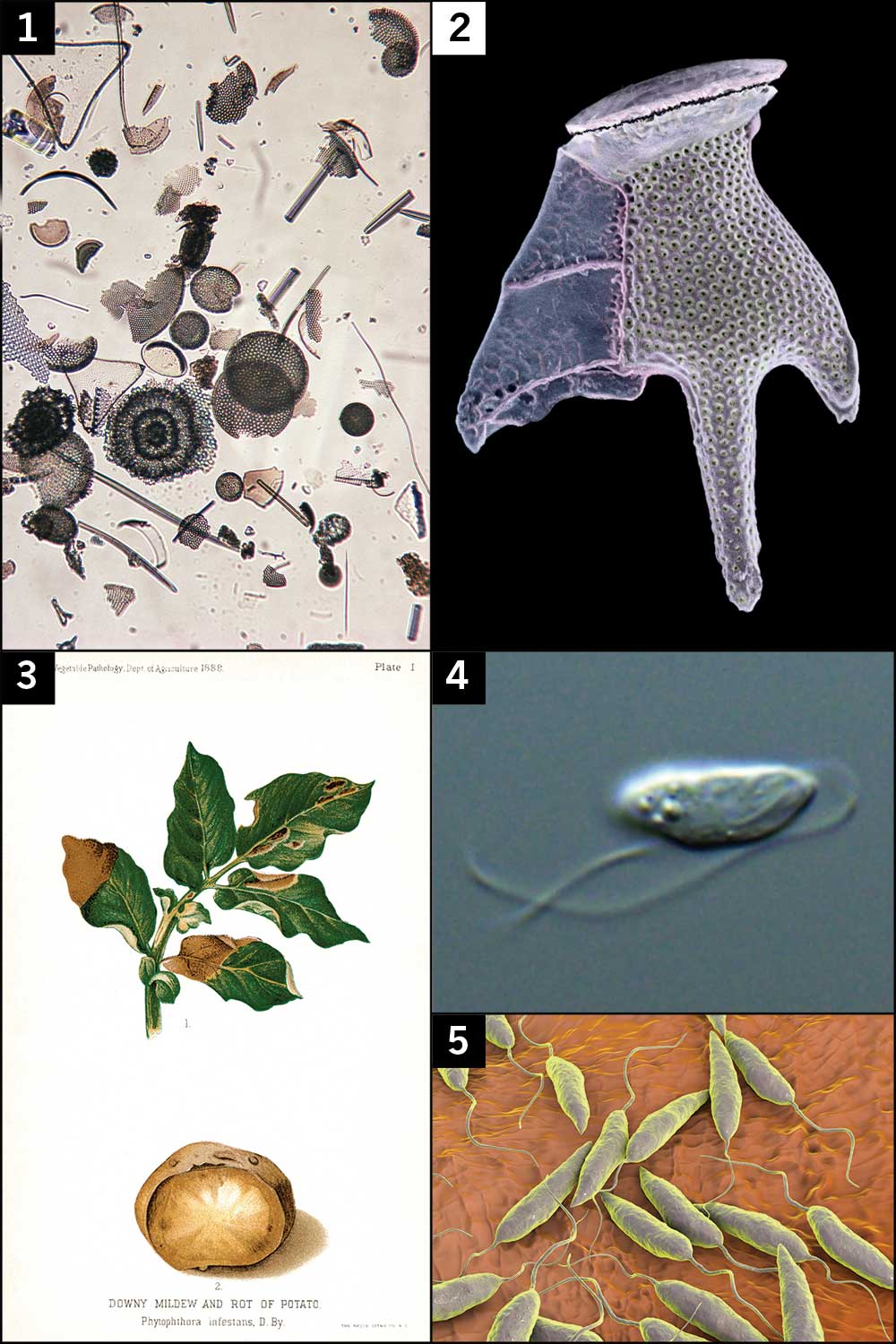
Getty; Minden Pictures/Alamy Stock Photo; Glasshouse Images/Alamy Stock Photo; Galina Prokopchuk and Daria Tashyreva; Science Photo Library/Alamy Stock Photo
But recent expeditions of the world’s oceans by the Tara schooner have raised their profile — not only are marine protists more abundant than expected, they are also extraordinarily more diverse. Tara plankton samples identified some 12,300 species of a single protist group, the diplonemids.
Protist diversity and their unique evolution limits our ability to understand their biology using standard animal models. When sequencing expressed eukaryote genes in samples taken by Tara, scientists realised that they had no idea what half of the genes do. In other words, they have no genetic equivalent in well-studied systems such as fruit flies or human cells.
One way that scientists can study the function of genes is by activating or deactivating the genes in the organisms to which they belong to see what happens. The technique is akin to turning a light switch on or off to figure out which bulb it is connected to. But such procedures require organisms that can be genetically manipulated — model organisms.
Without these systems, “we are blind to genetic information,” says Clarke.
Cutting and pasting
The Moore Foundation project has begun to fill the void. The first call for grant proposals was released in 2015, with the final tranche of funds distributed in 2018 to a total of 34 projects.
Nine teams were given two-year funding to apply precise and efficient gene editing techniques like CRISPR to more established systems such as diatoms. Another 25 teams were given one-year grants to trial techniques in untested species, with additional funds disbursed following annual progress assessments.
Despite the short window, the funds have spurred research in the field. Where previously only a few species of diatoms had been successfully manipulated, the project has led to genetic transformations in at least 12 species, six of which have passed down the changes to their progeny. Some of these species are the first to be genetically modified in their phylum — representing a group as large as plants and animals. The more established models have proven receptive to CRISPR and TALEN, another tool for cutting and repairing DNA.
For example, in 2018, Lukeš co-authored a paper reporting the first successful genetic manipulation of Diplonema papillatum, a species of the highly abundant and diverse diplonemids.
Less than 50 micrometres long, D. papillatum is shaped like a potato with two tiny roots, known as flagella. The researchers used an electroporation technique, which forms openings in the cell wall for objects to pass through, to introduce an antiobiotic resistance gene to the diplonemids. Organisms that had incorporated the gene into their DNA developed resistance to the bacteria — proof of a successful transformation. The entire process took three researchers a year to achieve. Having figured out how to introduce functional genes to the organisms, their next step is to improve the efficiency of the method.
Cell biologist, Ross Waller, and his team at the University of Cambridge, United Kingdom, have been able to add a protein to the cell wall of Perkinsus olseni, a parasite that infects oysters. While they weren’t the first to do it, they were able to take a closer look at the underlying mechanism of entry.
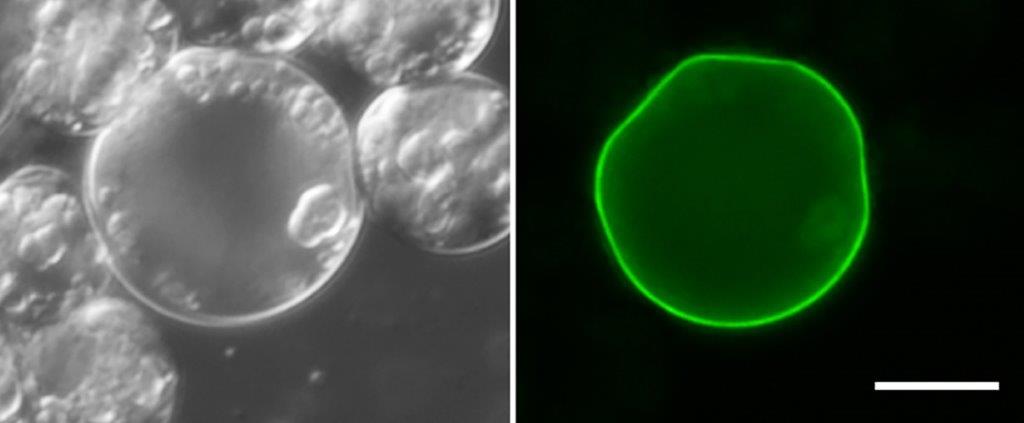
Adapted from W. F. Waller et al. PLoS Biology 16(7), e200633 (2018) via CC0 1.0
Their work on dinoflagellates has broken more ground. Dinoflagellates look like mini-stingrays and give corals their colour. Corals expel the dinoflagellates in times of stress, causing bleaching.
Waller’s team tried several methods to introduce genetic elements to the dinoflagellate nucleus, including infecting the cells with viruses, to no avail. But, in unpublished work on Amphidinium carterae, Waller and his colleagues have shown success in introducing DNA to the cell’s chloroplast, where dinoflagellates also store some of their genome. Previous efforts to introduce foreign elements to Amphidinium dinoflagellates could not be reproduced or created non-viable cell lines.
Other groups have shown only transient expression. Steven Wilhelm, a microbial ecologist at the University of Tennessee, Knoxville, for example, has introduced genes to a species that causes ecologically damaging brown algal blooms. The genes make Aureococcus anophagefferens fluoresce. But in Wilhelm’s samples, the glow only lasts for days, not months.
Open recipe
To speed up the pace of discovery, grantees were strongly encouraged by the Moore Foundation to exchange ideas and share progress. Moore facilitated these interactions through regular meetings, virtual workshops, and support for protocols.io, an open, online repository for scientific methods.
“They didn’t just award the grants and say ‘good luck’,” says Lenny Teytelman, co-founder and CEO of protocols.io, and a former yeast researcher.
At the Moore Foundation’s suggestion, the platform created a private group called Protist Research to Optimize Tools in Genetics (PROT-G), for researchers to upload and discuss their experimental methods.
Sharing protocols is not standard practice in the scientific community. Scientists who want to replicate a published study often have to contact the authors directly for a detailed how-to and hope for a response. A recent analysis of the biomedical literature published between 2015 and 2017 found that only one of the 104 articles with empirical data included a link to a complete protocol.
Since its launch in 2014, PROT-G has garnered 194 members, who have posted 169 public protocols, generating 53 discussions, and more than a thousand views just in October 2018. Additionally, 65 members have posted 214 private protocols, 149 of which have been shared with other PROT-G members.
The protocols are flagged as either working, in development, or unsuccessful. They include details about how to create artificial seawater, which brand of electroporation machines tolerate the saline solution, and the length and temperatures at which to incubate samples.
“Science has a rich history of not talking about what doesn’t work,” says Wilhelm, a grant recipient who was specifically tasked with promoting PROT-G within the community. “By sharing our failures, we have been able to help each other and avoid making the same mistakes over and over again,” he says.
Wilhelm initially worried about getting scooped in the open milieu. He was also apprehensive about investing in a project that wouldn’t lead to papers, setting back prospects for tenure and other grants. But he was quickly convinced of the benefits.
“There was a strong sense within the community, from early on, that we were participating in a global challenge with individual objectives,” says Waller in the UK. “It was a model for free sharing and engagement that most of us have not experienced in our research careers.”
With the final round of grants awarded this year, researchers are beginning to look elsewhere for funding. This time, though, they have more leverage to convince potential funders of research returns.
“To really push boundaries, we have to do risky things,” says Wilhelm.
This article is part of the Nature Index 360° series, which takes an all-round look at key topics in scientific research performance and publishing.
