Abstract
Previous epigenome-wide association studies (EWAS) of posttraumatic stress disorder (PTSD) and major depressive disorder (MDD) have been inconsistent. This may be due to small sample sizes, and measurement and tissue differences. The current two EWA analyses of 473 World Trade Center responders are the largest to date for both PTSD and MDD. These analyses investigated DNA methylation patterns and biological pathways influenced by differentially methylated genes associated with each disorder. Methylation was profiled on blood samples using Illumina 450 K Beadchip. Two EWA analyses compared current versus never PTSD, and current versus never MDD, adjusting for cell types and demographic confounders. Pathway and gene set enrichment analyses were performed to understand the complex biological systems of PTSD and MDD. No significant epigenome-wide associations were found for PTSD or MDD at an FDR P<0.05. The majority of genes with differential methylation at a suggestive threshold did not overlap between the two disorders. Pathways significant in PTSD included a regulator of synaptic plasticity, oxytocin signaling, cholinergic synapse and inflammatory disease pathways, while only phosphatidylinositol signaling and cell cycle pathways emerged in MDD. The failure of the current EWA analyses to detect significant epigenome-wide associations is in contrast with disparate findings from previous, smaller EWA and candidate gene studies of PTSD and MDD. Enriched gene sets involved in several biological pathways, including stress response, inflammation and physical health, were identified in PTSD, supporting the view that multiple genes play a role in this complex disorder.
Similar content being viewed by others
Introduction
The experience of trauma can have a pronounced negative impact on mental health, eliciting various forms of psychopathology. The most common sequelae of trauma are posttraumatic stress disorder (PTSD) and major depressive disorder (MDD).1, 2, 3, 4, 5 It is unclear how trauma elicits these disorders; however, recent evidence from animal models and human clinical studies suggests that changes to DNA methylation resulting from trauma may be one such mechanism.6, 7, 8, 9
Epigenetics of PTSD and depression
The majority of methylation studies of PTSD have focused on candidate genes selected on the basis of animal models studies (for example, glucocorticoid receptor (NR3C1;10, 11) or genetic association findings, such as FKBP5,12, 13 SLC6A4,14, 15 BDNF16 and ADCYAP1.17 Differential methylation regions within these candidate genes were also found to be associated with MDD (for example, refs 18, 19, 20, 21, 22), which is expected given their shared risk factors. Because knowledge about the etiology of PTSD and MDD is limited, the preferred method of explicating the genetic architectures underlying these disorders is the Epigenome Wide Association Study (EWAS) design. The EWAS design allows for a thorough investigation of the epigenetic patterns without relying on a priori knowledge of genetic risk factors.23, 24
PTSD EWAS findings
To date, there are three microarray-based EWAS examining epigenetic patterns in peripheral blood of cases with PTSD compared to trauma-exposed controls. First, Uddin et al.25 investigated methylation profiles of 100 participants (23 with lifetime PTSD). The study found that genes involved in the immune system were more likely to be un-methylated among individuals with PTSD. Second, Smith et al.26 found an overall increased level of methylation in PTSD in a sample of 110 participants (50 with current PTSD). The authors also reported differential methylation of five genes previously associated with inflammation (TPR, CLEC9A, ANAPC5, ANXA2 and TLR8). Third, Mehta et al.27 examined methylation profiles of 169 trauma-exposed participants (61 with current PTSD). PTSD was associated with methylation alterations that were in turn linked to gene expression differences in central nervous system development, apoptosis and growth rate networks. A comprehensive review of DNA methylation findings in PTSD can be found elsewhere.28, 29 Overall, the most promising results from human epigenetic studies of PTSD suggest methylation differences in immune function genes in cells of peripheral blood. However, the specific differentially methylated CpG sites identified in each study do not replicate across studies, possibly due to small sample sizes and methodological differences in cohort characteristics, types of trauma and platforms.
Depression EWAS findings
Candidate gene studies of MDD have reported differential methylation of specific genes including NR3C1, BDNF and SLC6A4 (for review see Januar et al.30). Seven EWAS of depression have been conducted. However, only two studies had samples of more than 100 participants: Nagy et al.31 analyzed methylation differences in post-mortem prefrontal cortex samples from 76 depressed individuals who committed suicide and 45 healthy controls. They found 115 differentially methylated regions, including regions related to astrocytic function, such as GRIK2 (glutamate receptor, ionotropic kainate 2) and BEGAIN (brain-enriched guanylate kinase-associated protein). Weder et al.32 conducted EWAS on saliva samples from 94 maltreated children and 94 controls and found that methylation in three genes—GRIN1, ID3 and TPPP—was correlated with depression symptoms. Their study also confirmed several candidate genes, including BDNF, NR3C1, and FKBP5. Among the 5 small N EWAS, 3 identified additional differentially methylated genes,33, 34, 35 although Sabunciyan et al. (2012)35 were unable to confirm their findings in an independent cohort; and 2 studies found global methylation level differences but no significant results for specific genes.36, 37 Importantly, EWA studies of MDD to date used a variety of tissue sources (post-mortem brain, saliva or blood), which might be one of the reasons, alongside differences in cohort characteristics, why differentially methylated CpG do not replicate across studies.
Limitations of PTSD and MDD EWAS
Overall, the EWAS approach has provided potentially promising insights into the pathophysiology of both PTSD and MDD, although to date, few if any results have been independently confirmed. Importantly, failure to replicate might also have occurred because EWA studies of depression have not controlled for exposure to extreme stressors, potent triggers of depression38, 39 (although Dempster et al.34 reported the same number of stressful life events in cases and controls). EWAS of PTSD and MDD also have not corrected for differences in the proportion of cell types within the tissue sample interrogated. This is particularly problematic in studies of whole blood, as specific regions of variable DNA methylation are responsible for defining cell lineage, and thus cell heterogeneity may act as a confounder when measuring DNA methylation in samples of peripheral blood without proper adjustment for differential cell counts.40, 41
Current study
Our study was designed to address the aforementioned limitations. We conducted two EWA analyses of DNA derived from peripheral blood to identify DNA methylation differences associated with PTSD and with MDD respectively, allowing us to test hypothesized genes (15 genes that emerged in previous literature are listed in Table 1) and investigate novel ones. We used the state-of-the-art 450k DNA methylation array and recruited a large sample (n=473) directly exposed to the 11 September 2001 World Trade Center (WTC) disaster. The current study contains the largest sample size to date for PTSD and MDD EWAS and to our knowledge, is the first to study methylation patterns that emerge for these conditions side by side. Finally, we used methylation findings to identify biological pathways that characterize PTSD and MDD. Cellular processes are regulated by a set of genes working in concert; thus biological pathway analysis has emerged as an important and perhaps more biologically valid approach for interpreting results from large-scale EWAS.
Materials and methods
Participants
Participants were recruited through the Stony Brook WTC-Health Program, part of a consortium of Clinical Centers of Excellence in the New York metropolitan area established in 2002 to monitor and treat WTC-related conditions in responders to the WTC disaster.42 Enrollees with documented WTC experience were enlisted from extensive outreach efforts involving partnerships with volunteer organizations, labor unions and public outlets. The current study was approved annually by the Committees on Research Involving Human Subjects at Stony Brook University (IRB number: 604113). Written informed consent was obtained.
The sample consisted of 473 responders assessed between February, 2012 and March, 2014. All participants provided blood samples for the epigenetics assays. Inclusion criteria were signed informed consent, sufficient English language skills to participate in a diagnostic interview, and being male. We included only males because females show notably different methylation patterns from males, and the cohort monitored at the Stony Brook WTC clinic is >90% male. We oversampled individuals with PTSD in order to have sufficient power for planned analyses. Participants were 49.5 years of age on average, predominantly Caucasian (>80%), and had similar rates of current PTSD and MDD (Table 2).
Clinical assessment
Master’s level clinical assessors were trained to administer the PTSD and MDD modules of the Structured Clinical Interview for DSM-IV (SCID43) with interval instructions (that is, worst episode of symptoms since 11 September 2001). SCID items were modified to assess PTSD symptoms in relation to traumatic WTC exposures (Criterion A). Before conducting the assessment, the interviewers reviewed participants’ occupational and medical histories in order to facilitate rapport and enhance the accuracy of interpretation of responses. Inter-rater agreement for 55 independently rated audio-tapes was very good (kappa≥0.82). Diagnoses were coded as (a) currently meets criteria for the disorder (current group), (b) met criteria since 11 September 2001 but does not meet currently (past group), and (c) did not meet criteria since 11 September 2001 (never group). Primary EWA analyses compared responders with current WTC-related PTSD to responders who never developed WTC-PTSD; these analyses were repeated for current MDD versus no MDD.
Illumina infinium human methylation 450K beadchip
Blood samples were obtained from each participant via venipuncture and sent to Roswell Park Cancer Institute for DNA extraction. Genomic DNA was isolated from 0.3 ml of whole blood using the Qiagen BioRobot Universal System and the QIAamp DNA blood BioRobot MDx Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s recommended protocol. DNA methylation profiling was performed by Roswell Park Cancer Institute using the Human Methylation 450 K BeadChip (Illumina, San Diego, CA, USA). DNA extraction and methylation profiling were done blinded to group assignment. Five hundred nanogram of high-quality genomic DNA measured by picogreen quantitation (Life technologies, Grand Island, NY, USA) was bisulphite converted, amplified, fragmented and hybridized to the Illumina Infinium Human Methylation 450 K Beadchip using standard Illumina protocol. Data were processed using Illumina's GenomeStudio methylation module (v1.9.0).
Data pre-processing and normalization
The 450K BeadChip methylation data from the GenomeStudio were imported into R (http://cran.r-project.org). The methylation data at each CpG probe were represented as a beta (β) values, that is, the ratio of methylated probe intensities to the total probe intensities. Pre-processing of methylation data at the 485 557 CpG probes was performed as follows. Probes with detection P-value >0.001 were set to missing, and probes with more than 20% missing were filtered. Beta mixture quantile (BMIQ) normalization44 was applied to the beta values for correction of bias due to the type I and type II probes. Non-specific, cross-hybridized probes,45, 46 probes overlapping with a single-nucleotide polymorphism, and probes mapping to repeat regions were filtered. The final data consisted of 375 223 CpG sites and 473 samples.
Estimation of blood cell type proportions
Cell type proportions have been implicated in DNA methylation analysis of whole blood samples.40, 47 The proportions of CD8T, CD4T, natural killer (NK), Bcell, monocytes (Mono) and granulocytes (Gran) were estimated using the R packages minfi and FlowSorted.Blood.450 based on the procedures described previously.40 We normalized the sum of the proportions per sample to one, and included five (CD8T, CD4T, NK, Bcell and Mono) out of six estimated cell types as an adjustment factor in our EWAS analysis. The association between each cell type and phenotype, adjusting for age, smoking status and race (Caucasian vs non-Caucasian), was carried out using linear regression.
Estimation of population stratification
Population stratification was estimated using the principal component approach based on the annotation file of autosomal CpG sites, which were within 10 bps of a variant identified in the 1000 Genome Project (Phase 1) with MAF>0.01.48
Statistical method for EWA analyses
To identify CpG sites associated with each phenotype, separate linear regression for each CpG was first fitted on logit transformed beta values (log(β/(1−β))) as response and diagnosis (current vs never), adjusting for age, smoking status, race, and cell types. We also compared the results by adjusting for first (1) 2 and (2) 10 estimated population stratification principal components instead of race in our model. The logit transformed β values, also known as the M-values, had been recommended for conducting differential methylation analysis.49, 50 A false discovery rate (FDR)51 was used to account for multiple testing. Post hoc analysis was carried out to examine whether the past PTSD (MDD) group show differential methylation patterns on candidate CpG sites in comparison to methylation values of current and never groups. Principal component method was used to reduce the multiple candidate CpG sites into a single eigen CpG. The P-values for the post hoc comparison were computed from two-tailed two sample t-tests. Global methylation level was defined as the average methylation (beta values averaged over all the CpG sites). We also considered the average methylation defined by the beta values averaged over the 22 622 CpG sites which overlapped with the Infinium HumanMethylation27 Beadchip. The P-values for comparison of global methylation level were computed from two-tailed two sample t-tests. The effect of WTC exposures3 on the EWA analyses were also examined (Supplementary Materials), but did not change the results in a way that would alter the interpretation.
Pathway and gene ontology analyses
Pathway and gene ontology analyses were carried out using the gometh function in the Bioconductor package missMethyl.52 Since the number of CpG sites mapping to each gene varied in the Methylation 450 K BeadChip, pathway and gene ontology analysis would be biased and inaccurate.53 gometh accounted for the varying number of CpG sites per gene by providing a prior probability for each gene based on gene length, followed by a modified hypergeometric test for over-representation of a gene set.54 We tested for over-representation among the top 100 to 500 (in increment of 50) CpG sites for each analysis, against the background list of 375 223 CpG sites. In all, 5776 gene ontologies including biological processes, molecular functions and cellular components, and 290 KEGG pathways (minimum and maximum number of genes for each gene set were 15 and 500, respectively) were tested. Overlapping gene sets significant at FDR 0.05 for all the top K CpG sites were reported.
Data availability
The methylation data is available at the Psychiatric Genomics Consortium (PGC) website https://www.med.unc.edu/pgc/.
Results
EWAS analyses
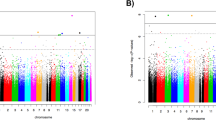
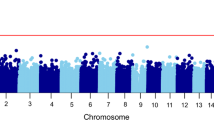
EWAS analyses with each diagnostic group did not identify statistically significant CpG sites at FDR 0.05. The list of CpG sites at a nominal P-value 0.0001 that did not reach a significant FDR cutoff is provided in Table 3. The signs of the estimated coefficients remain consistent within the subset analysis, that is, PTSD case vs control within never MDD and MDD case vs control within current PTSD. The top suggestive CpG for PTSD was cg05693864 at gene body of ZDHHC11, with the mean methylation difference between current and never groups of 2.7%. The top suggestive CpG for MDD was cg19722082 on EDIL3 gene (−2.5% mean methylation difference). Only three CpG sites common to both PTSD and MDD emerged at the suggestive level. The most prominent of them was cg06182923, located on the gene body of CSMD2 (−3.1% in PTSD and −3.5% in MDD). Due to the high correlation between the estimated cell type proportions and the population stratification principal components, our primary analyses were based on a model which adjusted for age, smoking status, race and cell types (Supplementary Table 1). Additional details on the secondary analyses based on a model which adjusted for population stratification principal components were provided in Supplementary Materials.
Principal component analyses were performed on the list of CpG sites at nominal P-value 0.0001 for each of PTSD and MDD. The methylation profiles were represented using the first principal component, that is, the eigen CpG. Figure 1 compares the eigen CpG for current, past and never groups of PTSD and MDD. The past PTSD group exhibited intermediate eigen CpG values (P=1.12e−05 for current vs past, P=0.00331 for past vs never), suggesting that the overall methylation profiles of the top ranking CpG sites were associated with disease progression. For MDD, the past group was not statistically different from the never group (P=0.0902), whereas the current was statistically different from the past group (P=0.00915).
Of note, none of CpG sites mapping on the previously identified candidate genes (FKBP5, NR3C1, BDNF, SLC6A4, ADCYAP1, TPR, CLEC9A, ANAPC5, ANXA2, TLR8, GRIK2, BEGAIN, GRIN1, ID3 and TPPP) was significant at nominal P-value.0001. To evaluate any trends in these genes, we examined the direction of methylation on CpG sites that exhibited differential methylation between current and never groups at nominal P-value⩽0.05 (Table 1). These CpG sites were mostly hypomethylated in PTSD and MDD. One study26 observed a higher average methylation level in PTSD relative to non-PTSD based on the Infinium HumanMethylation27 Beadchip. However, our global methylation pattern (defined using either all CpG sites or CpG sites which overlapped with the Infinium HumanMethylation27 Beadchip) did not show a statistically significant difference (P-value=0.589 and 0.884, respectively) comparing current PTSD to control. The P-values were 0.911 (all CpG sites) and 0.942 (Infinium HumanMethylation27 CpG sites) comparing current MDD to control.
Pathway and gene ontology analyses
Twenty-three KEGG pathways were significant at FDR 0.05 for PTSD, and two were significant for MDD (Table 4). The top KEGG pathways that emerged for PTSD included cGMP-PKG, oxytocin and MAPK signaling, insulin resistance, cholinergic synapse and inflammatory bowel disease pathways. The two KEGG pathways that emerged for MDD are implicated in phosphatidylinositol signaling system and cell cycle. No gene ontologies were enriched at FDR 0.05 for both PTSD and MDD. The results of the cell type proportion analyses are presented in Supplementary Figure S1. In addition, both the results from EWAS and pathway analyses remained consistent after taking into account potential confounding due to exposure (Supplementary Materials).
Discussion
The current study reports results from the largest EWA analyses of PTSD and of MDD to date, investigating DNA methylation differences underpinning these two conditions. We found no significant genome-wide methylation differences between responders with current versus no WTC-related disorder, suggesting that links between individual methylation sites and these conditions are too subtle to be detected in such a sample. However, we identified disorder-specific biological pathways underpinned by differentially methylated genes. PTSD was associated with pathways regulating neuron signaling, inflammation and multiple aspects of physical health. The current study makes an important contribution to the understanding of methylation in PTSD and in MDD, evaluating hypotheses that emerged in research on these phenotypes.
EWAS findings
Our null findings are in contrast to previously published EWAS results of PTSD and depression. Using a much larger sample than previous studies, we did not confirm the genes implicated in prior studies of DNA methylation in PTSD or MDD. Previous work has suggested that differentially methylated sites were significantly overrepresented in immune system genes for both disorders. However, no individual immune system gene was identified in this study. Also, we did not observe statistically significant methylation differences in 15 candidate genes identified by prior studies. Furthermore, analyses of trends suggested hypomethylation of FKBP5, NR3C1, BDNF and SLC6A4 in PTSD and MDD, whereas prior studies found hypermethylation in NR3C1, BDNF and SLC6A4,15, 30, 55, 56 raising questions about their role in the methylation signature of PTSD and MDD.
Methodological differences between our study and previous EWA studies might account for some of the discrepancies in the results, and as such our findings need to be considered in the context of the current sample characteristics (especially gender composition) as well as methodology. First, our study focused on men, while previous studies included more women than men. Growing literature demonstrates that DNA methylation changes occur in a sex-specific manner.57 Second, previous PTSD EWA studies were performed on populations that were predominantly of African American ancestry, while our sample is mostly of European American ancestry. Recent reports have found significant differences in DNA methylation between African American and Caucasian ancestry subjects. This suggests that ancestry-specific genetic background may play a role in shaping the epigenetic landscape of the human genome in ways similar to SNP-GWAS population stratification.48 Third, trauma exposure in our study was a specific event common to all responders—the WTC disaster. Previous studies considered retrospectively reported traumas that were not common to all individuals and were uncorroborated.58, 59, 60
Fourth, previous EWAS of MDD used a variety of tissue sources, with only one study33 using blood. Fifth, previous EWAS of MDD did not consider trauma-exposed populations, which might be a subgroup characterized by somewhat different depression etiology.58, 59, 60 Finally, prior studies did not control for blood cell types. This is particularly problematic because findings in PTSD of differential methylation amongst genes involved in immune response could represent greater number of immune cells in the blood sample. Controlling for differences in the number of immune cells likely affects the ability to detect these differences. Overall, each study to date, including the current study, applied different methodologies and thus cannot be considered as direct replications of one another.
Biological pathways
We also conducted pathway analyses in the current sample in order to identify significantly enriched gene sets associated with PTSD and MDD. PTSD was characterized by a wide range of pathways, many of which map onto the biological mechanisms implicated by previous methylation studies of PTSD. First, the cGMP-PKG signaling pathway has been identified as the top KEGG pathway for PTSD, which was shown to regulate synaptic plasticity and fear memory consolidation in the lateral amygdala.61 The second most significant pathway for PTSD was oxytocin signaling. Neuropeptide oxytocin has been shown to be an important anti-stress factor of the brain62 and implicated in PTSD.63, 64 Third, we identified several pathways related to immunity (for example, inflammatory bowel disease and rheumatoid arthritis pathways), which is in line with a large body of evidence that PTSD is associated with altered inflammatory processes.65, 66, 67 Several genes (most notably FKBP5) are thought to play a role in regulation of the HPA axis and immunological responses to stress.13, 68, 69 Although the current study has not confirmed differential methylation of these individual genes, the observed immunity-related pathways are consistent with previous research. Fourth, several cancer-related pathways (for example, MAPK signaling pathway), as well as pathways related to cardiovascular and metabolic disease (for example, insulin resistance pathway) and the nervous system (for example, cholinergic synapse pathway) emerged for PTSD, which are in line with the considerable co-occurrence of PTSD with a range of physical disorders.3, 70, 71, 72
Conversely, only two pathways emerged for MDD. First, phosphatidylinositol signaling system was identified as the top significant KEGG pathway. The phosphatidylinositol signaling system has broad physiological significance, and has been shown to be affected by antidepressants in several studies.73, 74, 75, 76 The second pathway significant in MDD was cell cycle, a broad pathway without specific links to depression. It is possible that the smaller number of MDD cases resulted in lower power to detect significant MDD pathways.
Limitations
The current study had several strengths, including the largest EWAS sample to date in research on PTSD and MDD, and exposure to a common traumatic even for all participants. Nonetheless, our findings must be considered in the context of several limitations. First, since our study is cross-sectional, we cannot determine whether observed alterations in the epigenome of PTSD affected patients are a consequence of the disease or a part of its etiology. By identifying a trauma-exposed, unaffected comparison group, we guarded against differential methylation being just a consequence of trauma exposure. Second, the sample size of our study is relatively small for EWAS, even if it is the largest among PTSD and MDD methylation studies.77, 78 Third, our methylation analysis was performed in DNA samples derived from peripheral blood cells and were thus a mix of cell types. We sought to control for the mix statistically, which emerged as the state-of-the-art method, but future work needs to isolate and examine each cell type individually. Fourth, as discussed above, sample characteristics, tissue used and other methodological aspects of the current study differed substantially from previous EWAS of PTSD and MDD. In particular, we focused on males while prior studies relied on predominantly female samples. Larger samples are needed to investigate the impact of gender differences on methylation in PTSD, an important open question. Future studies designed as direct replications of prior research need to be conducted, ideally using large sample sizes, to confirm previous findings. Finally, a stronger association between methylation and diagnoses could potentially be derived by considering allele specific methylation as observed in Klengel et al.13 or by also considering lifetime disorders not associated with 9/11.
Conclusions
The current study aimed to provide a better understanding of the relationship between epigenetic alteration and PTSD and MDD. We found no epigenome-wide significant hits for either condition, and as such did not confirm findings from previous, smaller EWAS and candidate gene studies of PTSD and MDD. Nonetheless, we identified enriched gene sets involved in several biological pathways associated with PTSD, including stress response, inflammation, neural signaling and physical health.
References
Smith TC, Ryan MA, Wingard DL, Slymen DJ, Sallis JF, Kritz-Silverstein D et al. New onset and persistent symptoms of post-traumatic stress disorder self reported after deployment and combat exposures: prospective population based US military cohort study. BMJ 2008; 336: 366–371.
Wells TS, LeardMann CA, Fortuna SO, Smith B, Smith TC, Ryan MA et al. A prospective study of depression following combat deployment in support of the wars in Iraq and Afghanistan. Am J Public Health. 2010; 100: 90–99.
Wisnivesky JP, Teitelbaum SL, Todd AC, Boffetta P, Crane M, Crowley L et al. Persistence of multiple illnesses in World Trade Center rescue and recovery workers: a cohort study. Lancet 2011; 378: 888–897.
Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE . Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 2005; 62: 593–602.
O'Donnell ML, Creamer M, Pattison P . Posttraumatic stress disorder and depression following trauma: understanding comorbidity. Am J Psychiatry. 2004; 161: 1390–1396.
Heinzelmann M, Gill J . Epigenetic mechanisms shape the biological response to trauma and risk for PTSD: a critical review. Nurs Res Pract 2013; 2013: 417010.
Schmidt U, Kaltwasser SF, Wotjak CT . Biomarkers in posttraumatic stress disorder: Overview and implications for future research. Dis Markers 2013; 35: 43–54.
Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR et al. Epigenetic programming by maternal behavior. Nat Neurosci 2004; 7: 847–854.
Kinnally EL, Capitanio JP, Leibel R, Deng L, LeDuc C, Haghighi F et al. Epigenetic regulation of serotonin transporter expression and behavior in infant rhesus macaques. Genes Brain Behav 2010; 9: 575–582.
McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 2009; 12: 342–348.
Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F et al. Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans with posttraumatic stress disorder. Biol Psychiatry 2015; 77: 356–364.
Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB et al. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA 2008; 299: 1291–1305.
Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci 2013; 16: 33–41.
Chang S-C, Koenen KC, Galea S, Aiello AE, Soliven R, Wildman DE et al. Molecular variation at the SLC6A3 locus predicts lifetime risk of PTSD in the Detroit Neighborhood Health Study. PLoS ONE 2012; 7: e39184.
Koenen KC, Uddin M, Chang SC, Aiello AE, Wildman DE, Goldmann E et al. SLC6A4 methylation modifies the effect of the number of traumatic events on risk for posttraumatic stress disorder. Depress Anxiety 2011; 28: 639–647.
Roth TL, Lubin FD, Funk AJ, Sweatt JD . Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry 2009; 65: 760–769.
Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature 2011; 470: 492–497.
Na K-S, Chang HS, Won E, Han K-M, Choi S, Tae WS et al. Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS ONE 2014; 9: e85425.
Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM . Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 2008; 3: 97–106.
Olsson C, Foley D, Parkinson-Bates M, Byrnes G, McKenzie M, Patton G et al. Prospects for epigenetic research within cohort studies of psychological disorder: a pilot investigation of a peripheral cell marker of epigenetic risk for depression. Biol Psychol 2010; 83: 159–165.
Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A . The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet 2008; 147: 543–549.
Smart C, Strathdee G, Watson S, Murgatroyd C, McAllister-Williams R . Early life trauma, depression and the glucocorticoid receptor gene–an epigenetic perspective. Psychol Med 2015; 45: 3393–3410.
Koenen KC, Duncan LE, Liberzon I, Ressler KJ . From candidate genes to genome-wide association: The challenges and promise of posttraumatic stress disorder genetic studies. Biol Psychiatry 2013; 74: 634.
Rakyan VK, Down TA, Balding DJ, Beck S . Epigenome-wide association studies for common human diseases. Nat Rev Genet 2011; 12: 529–541.
Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci USA 2010; 107: 9470–9475.
Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B et al. Differential immune system DNA methylation and cytokine regulation in post‐traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet 2011; 156: 700–708.
Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW et al. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci USA 2013; 110: 8302–8307.
Vinkers CH, Kalafateli AL, Rutten BP, Kas MJ, Kaminsky Z, Turner JD et al. Traumatic stress and human DNA methylation: a critical review. Epigenomics 2015; 7: 593–608.
Zannas AS, Provençal N, Binder EB . Epigenetics of posttraumatic stress disorder: current evidence, challenges, and future directions. Biol Psychiatry 2015; 78: 327–335.
Januar V, Saffery R, Ryan J . Epigenetics and depressive disorders: a review of current progress and future directions. Int J Epidemiol 2015; 44: 1364–1387, dyu273.
Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C et al. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol Psychiatry 2015; 20: 320–328.
Weder N, Zhang H, Jensen K, Yang BZ, Simen A, Jackowski A et al. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J Am Acad Child Adolesc Psychiatry 2014; 53: 417–424, e5.
Davies MN, Krause L, Bell JT, Gao F, Ward KJ, Wu H et al. Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biol 2014; 15: 1.
Dempster EL, Wong CC, Lester KJ, Burrage J, Gregory AM, Mill J et al. Genome-wide methylomic analysis of monozygotic twins discordant for adolescent depression. Biol Psychiatry 2014; 76: 977–983.
Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE et al. Genome-wide DNA methylation scan in major depressive disorder. PLoS ONE 2012; 7: e34451.
Byrne E, Carrillo-Roa T, Henders A, Bowdler L, McRae A, Heath A et al. Monozygotic twins affected with major depressive disorder have greater variance in methylation than their unaffected co-twin. Transl Psychiatry 2013; 3: e269.
Uddin M, Koenen K, Aiello AE, Wildman D, de Los Santos R, Galea S . Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol Med 2011; 41: 997–1007.
Hammen C . Stress and depression. Annu Rev Clin Psychol. 2005; 1: 293–319.
Monroe SM, Simons AD . Diathesis-stress theories in the context of life stress research: implications for the depressive disorders. Psychol Bull 1991; 110: 406–425.
Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012; 13: 1.
Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén S-E, Greco D et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE 2012; 7: e41361.
Herbert R, Moline J, Skloot G, Metzger K, Baron S, Luft B et al. The World Trade Center disaster and the health of workers: five-year assessment of a unique medical screening program. Environ Health Perspect 2006; 1853–1858.
First MB, Spitzer RL, Gibbon M, Williams JB . Structured Clinical Interview for DSM-IV Axis I Disorders: Patient Edition (February 1996 Final), SCID-I/P: Biometrics Research Department. New York State Psychiatric Institute: New York, NY, 1998.
Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013; 29: 189–196.
Chen Y-a, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013; 8: 203–209.
Price EM, Cotton AM, Lam LL, Farré P, Emberly E, Brown CJ et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenet Chromatin 2013; 6: 1.
Houseman EA, Kim S, Kelsey KT, Wiencke JK . DNA methylation in whole blood: uses and challenges. Curr Environ Health Rep 2015; 2: 145–154.
Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R et al. Accounting for population stratification in DNA methylation studies. Genet Epidemiol 2014; 38: 231–241.
Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010; 11: 587.
Kuan PF, Wang S, Zhou X, Chu H . A statistical framework for Illumina DNA methylation arrays. Bioinformatics 2010; 26: 2849–2855.
Benjamini Y, Hochberg Y . Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995; 51: 289–300.
Phipson B, Maksimovic J, Oshlack A . missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics 2016; 32: 286–288.
Geeleher P, Hartnett L, Egan LJ, Golden A, Raja Ali RA, Seoighe C . Gene-set analysis is severely biased when applied to genome-wide methylation data. Bioinformatics 2013; 29: 1851–1857.
Young MD, Wakefield MJ, Smyth GK, Oshlack A . Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010; 11: R14.
Kang H-J, Kim J-M, Stewart R, Kim S-Y, Bae K-Y, Kim S-W et al. Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Progress Neuropsychopharmacol Biol Psychiatry 2013; 44: 23–28.
Perroud N, Paoloni-Giacobino A, Prada P, Olié E, Salzmann A, Nicastro R et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl Psychiatry 2011; 1: e59.
Uddin M, Sipahi L, Li J, Koenen KC . Sex differences in DNA methylation may contribute to risk of PTSD and depression: a review of existing evidence. Depress Anxiety 2013; 30: 1151–1160.
Harald B, Gordon P . Meta-review of depressive subtyping models. J Affect Disord 2012; 139: 126–140.
Heim C, Mletzko T, Purselle D, Musselman DL, Nemeroff CB . The dexamethasone/corticotropin-releasing factor test in men with major depression: role of childhood trauma. Biol Psychiatry. 2008; 63: 398–405.
Heim C, Newport DJ, Mletzko T, Miller AH, Nemeroff CB . The link between childhood trauma and depression: insights from HPA axis studies in humans. Psychoneuroendocrinology 2008; 33: 693–710.
Ota KT, Pierre VJ, Ploski JE, Queen K, Schafe GE . The NO-cGMP-PKG signaling pathway regulates synaptic plasticity and fear memory consolidation in the lateral amygdala via activation of ERK/MAP kinase. Learn Mem 2008; 15: 792–805.
Neumann ID, Slattery DA . Oxytocin in general anxiety and social fear: a translational approach. Biol Psychiatry. 2016; 79: 213–221.
Eidelman-Rothman M, Goldstein A, Levy J, Weisman O, Schneiderman I, Mankuta D et al. Oxytocin affects spontaneous neural oscillations in trauma-exposed war veterans. Front Behav Neurosci 2015; 9: 165.
Seng JS . Posttraumatic oxytocin dysregulation: is it a link among posttraumatic self disorders, posttraumatic stress disorder, and pelvic visceral dysregulation conditions in women? J Trauma Dissoc 2010; 11: 387–406.
Michopoulos V, Powers A, Gillespie CF, Ressler KJ, Jovanovic T . Inflammation in fear- and anxiety-based disorders: PTSD, GAD, and beyond. Neuropsychopharmacology 2017; 42: 254–270.
Passos IC, Vasconcelos-Moreno MP, Costa LG, Kunz M, Brietzke E, Quevedo J et al. Inflammatory markers in post-traumatic stress disorder: a systematic review, meta-analysis, and meta-regression. Lancet Psychiatry 2015; 2: 1002–1012.
Rosen RL, Levy-Carrick N, Reibman J, Xu N, Shao Y, Liu M et al. Elevated C-reactive protein and posttraumatic stress pathology among survivors of the 9/11 World Trade Center attacks. J Psychiatr Res. 2017; 89: 14–21.
Mahon PB, Zandi PP, Potash JB, Nestadt G, Wand GS . Genetic association of FKBP5 and CRHR1 with cortisol response to acute psychosocial stress in healthy adults. Psychopharmacology (Berl) 2013; 227: 231–241.
Yehuda R, Cai G, Golier JA, Sarapas C, Galea S, Ising M et al. Gene expression patterns associated with posttraumatic stress disorder following exposure to the World Trade Center attacks. Biol Psychiatry 2009; 66: 708–711.
Scott KM, Lim C, Al-Hamzawi A, Alonso J, Bruffaerts R, Caldas-de-Almeida JM et al. Association of mental disorders with subsequent chronic physical conditions: world mental health surveys from 17 countries. JAMA Psychiatry 2016; 73: 150–158.
Swartzman S, Booth JN, Munro A, Sani F . Posttraumatic stress disorder after cancer diagnosis in adults: a meta-analysis. Depress Anxiety. 2016; 34: 327–339.
Ahmadi N, Arora R, Vaidya N, Yehuda R, Ebrahimi R . Post-traumatic stress disorder is associated with increased incidence of insulin resistance and metabolic syndrome. J Am Coll Cardiol 2013; 61: E1347.
Donati RJ, Rasenick MM . G protein signaling and the molecular basis of antidepressant action. Life Sci. 2003; 73: 1–17.
Harwood AJ . Lithium and bipolar mood disorder: the inositol-depletion hypothesis revisited. Mol Psychiatry. 2005; 10: 117–126.
Racagni G, Popoli M . Cellular and molecular mechanisms in the long-term action of antidepressants. Dialogues Clin Neurosci 2008; 10: 385–400.
Xu X, Muller-Taubenberger A, Adley KE, Pawolleck N, Lee VW, Wiedemann C et al. Attenuation of phospholipid signaling provides a novel mechanism for the action of valproic acid. Eukaryot Cell. 2007; 6: 899–906.
Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry 2014; 71: 255–264.
Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol 2013; 31: 142–147.
Acknowledgements
The study was funded by CDC/NIOSH award U01 OH010416-01 (PI: Benjamin J. Luft). We gratefully acknowledge the support of the first responders and rescue/recovery workers for participating in this study. We also thank the staff of the Stony Brook World Trade Center Health Program and the World Trade Center Health Program Data Monitoring Center for ongoing support. We are also grateful to Sean Glenn at the Roswell Park Cancer Institute for his contributions to the methylation assessment. We also thank Sandro Galea (Boston University), Steve Horvath (UCLA), Andrew Ratanatharathorn (Columbia University), and Sean Clouston (Stony Brook University) for their invaluable input on the project. The findings and conclusions in this article are those of the authors and do not represent the official position of NIOSH, the CDC or the US Public Health Service. The analysis was supported by U01 OH010416-01 (PI: Benjamin J. Luft) from Centers for Disease Control and Prevention (CDC) and The National Institute for Occupational Safety and Health (NIOSH).
Disclaimer
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Kuan, PF., Waszczuk, M., Kotov, R. et al. An epigenome-wide DNA methylation study of PTSD and depression in World Trade Center responders. Transl Psychiatry 7, e1158 (2017). https://doi.org/10.1038/tp.2017.130
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2017.130
This article is cited by
-
Epigenetics of early-life adversity in youth: cross-sectional and longitudinal associations
Clinical Epigenetics (2022)
-
Meta-analysis of epigenome-wide association studies of major depressive disorder
Scientific Reports (2022)
-
Long non-coding RNA LINC00926 regulates WNT10B signaling pathway thereby altering inflammatory gene expression in PTSD
Translational Psychiatry (2022)
-
Epigenome-wide association study of posttraumatic stress disorder identifies novel loci in U.S. military veterans
Translational Psychiatry (2022)
-
Epigenome-wide meta-analysis of PTSD symptom severity in three military cohorts implicates DNA methylation changes in genes involved in immune system and oxidative stress
Molecular Psychiatry (2022)