Abstract
A cold-induced transcript encoding a Casparian strip membrane domain (CASP)-like protein (ClCASPL) was identified in watermelon (Citrullus lanatus). Fluorescence microscopy analysis showed that ClCASPL-GFP is localized in the plasma membrane. The orthologous gene in Arabidopsis thaliana (AtCASPL4C1) was also found to play an important role in cold tolerance. Expression analysis using a β-glucuronidase (GUS) reporter reveals that AtCASPL4C1 is widely expressed in a variety of organs and is cold inducible. Analysis of AtCASPL4C1 T-DNA knock-out plants showed altered growth dynamics, faster growth, increased biomass (dry weight) and earlier flowering compared to wild type (Col-0) and ClCASPL overexpressing plants. AtCASPL4C1 knock-out plants showed elevated tolerance to cold stress, while overexpressing CICASPL resulted in increased sensitivity to cold stress in Arabidopsis. Interestingly, AtCASPL4C1 knock-out plants did not display significant alterations in the Casparian strip formation in roots. Thus, the combination of these results suggests a role for CICASPL and AtCASPL4C1 beyond Casparian strip formation in roots, possibly indicating a more fundamental role in vascular tissue.
Similar content being viewed by others
Introduction
Abiotic stresses are major environmental factors that adversely affect plant growth, development and yield. Plants have evolved complex signaling networks to adapt to abiotic stresses via modulating various physiological and biochemical processes1,2,3. These stress signals are perceived by receptors, transduced and propagated by downstream effectors, ultimately altering the expression of a variety of genes that determine growth, tolerance and/or survival depending on the severity of the environmental conditions2,4.
Transmembrane (TM) proteins located in the plasma membrane are known to have diverse physiological functions including signal perception and recognition, via ion and metabolite exchange. In Arabidopsis, approximately 6,500 proteins are predicted to be TM proteins5. Some integral membrane proteins are induced by stress, such as drought stress in Triticum turgidum6, or salt stress in Xerophyta viscose7 as well as having a role during development, e.g. kernel development in Hordeum vulgare8.
The Casparian strip, first described by Robert Caspary in 18659, is a ring-like cell wall structure in the root endodermis of vascular plants. The role of the Casparian strip is to block the passive flow of materials in vascular plants10. Recent reports showed that the Casparian strip is composed of a lignin polymer without suberin in Arabidopsis11. This lignin polymer structure generates a para-cellular barrier, analogous to tight junctions in animals, that is thought to be crucial for selective nutrient uptake, exclusion of pathogens and many other processes10. Casparian strip membrane domain proteins (CASP, CASP1/2/3/4/5) are crucial for Casparian strip formation in the endodermis in plants. CASP1/2/3/4/5 belong to ‘uncharacterized protein family’ UPF0497 (39 members) in Arabidopsis9. These CASPs are proposed to form an extensive, transmembrane polymeric platform and were speculated to guide the assembly and activity of lignin biosynthetic enzymes9. Recently, evolutionary analysis of CASP family genes indicated that CASPL genes belong to the MARVEL (MAL and related proteins for vesicle trafficking and membrane link) protein family, which has only been experimentally described in metazoans, to date12.
Previously, we identified a gene encoding a cold-induced integral membrane protein in watermelon13. The orthologue to this gene was identified to be At3g55390 in Arabidopsis thaliana, where it is annotated as the CASP-LIKE PROTEIN 4C1. At3g55390 belongs to the CASP protein family containing the five CASP genes (CASP1/2/3/4/5), which are known to mediate Casparian strip formation in plants9. Thereafter, these are referred to as ClCASPL (from watermelon) and AtCASPL4C1 (CASP-like) or AtCASPL4C1 from Arabidopsis, respectively. In this study, we investigated the role of ClCASPL and AtCASPL4C1 in growth and cold tolerance in watermelon and Arabidopsis, respectively.
Materials and Methods
Plant Materials and treatment
An IVSM-9 inbred line of watermelon (Citrullus lanatus) and wild type (WT) of Arabidopsis (Arabidopsis thaliana) (Col-0) were used for amplification of ClCASPL and AtCASPL4C1 (At3G55390) genes. WT Arabidopsis thaliana (Col-0) was used for transformation to construct OX-ClCASPL of Arabidopsis. The SALK_034800C line was used for screening of homozygous Atcaspl4c1 knock-out mutant plants. 3-week-old plants in Jiffy seedling culture substrate or 2-week-old seedlings cultured in 1/2 MS medium of WT, Atcaspl4c1 and OX-ClCASPL Arabidopsis were used for analysis of cold stress and phenotypes evaluations. Tobacco (Nicotianaben thamiana) was used for transient expression of the ClCASPL gene.
5 days old plants were transferred onto half-strength MS (Murashige-Skoog, sigma-Aldrich) medium and grew under 10 °C, light/dark (16 h/8 h) conditions. For soil growth plants, 21 days old plants were used to cold treatments, under 10 °C, light/dark (16 h/8 h) conditions. The pictures and data were collected at the indicated time. The values are means ± SD (n = 20). Bar = 1 cm. Star signs indicate a significantly difference (p < 0.05, student’s t test).
Phylogenetic Tree Construction
The ClustalW program was used for alignment of ClCASPL with the AtCASPL4C1 protein sequence, which was obtained from the TAIR database. After alignment by ClustalW, a Neighbour-Joining tree was constructed by using MEGA 6.0, with 1000 as the number of bootstrap replications. The 39 members of CASP family genes were included from TAIR database. The TMHMM program was used for transmembrane region identification for ClCASPL and AtCASPL4C1.
Transient Expression of ClCASPL Gene in Tobacco
The amplification of ClCASPL coding sequence without a termination codon was linked to pMDC83 binary expression vector to generate the ClCASPL-pMDC83 by Gateway cloning14. The ClCASPL-GFP plasmid was introduced into Agrobacterium tumefaciens strain (GV1301). Leaves from tobacco (Nicotianaben thamiana) were used for transformation and for checking transient expression according to the procedures15. Various organelle-targeted markers fused with RFP were used as controls to determine sub-cellular localization16. These tobacco leaves were observed using a confocal laser scanning microscope (ZEISS). The primers used in this study for GFP fusion are listed in Supplementary Table 1.
Expression of CASPL
In silico transcript abundance analysis for genes encoding proteins of the CASP family was carried out as outlined in Narsai et al.17. The promoter sequences were searched by A Database of Plant Promoter Sequences (http://linux1.softberry.com/berry.phtml?topic=plantprom&group=data&subgroup=plantprom). The promoter region of At3g55390 was amplified and inserted into a region upstream of the GUS gene of within the pMDC162 binary expression vector using the gateway system14. The resulting construct was transformed in the Agrobacterium tumefaciens strain GV3101. Transformation of Arabidopsis was conducted according to the floral-dip method18. Tissues from transgenic plants were collected in microcentrifuge tubes. Subsequently, the samples were stained as previously published procedures19.
Cold-induced expression of CASPL was analyzed using qPCR and CASPL-promoter analysis using ClCASPL promoter-GUS activity after cold stress. GUS activity was measured as previously published procedures20. The primers used for promoter amplification are listed in Supplementary Table 1.
AtCASPL4C1 and OX-ClCASPL Construction
cDNA amplification of the ClCASPL gene was inserted to pMDC32 binary expression vector to generate the ClCASPL-pMDC32 by gateway approaches14. The ClCASPL-pMDC32 plasmid was introduced into Agrobacterium tumefaciens strain (GV1301). The floral dipping method was used to generate OX-ClCASPL of Arabidopsis18. For screening of transgenic plants, seeds harvested from transformed Arabidopsis plants were surface-sterilized and plated onto MS medium containing 50 mg/L hygromycin B. The plated seeds were vernalized at 4 °C for 2–4 days in the dark to synchronize germination and then transferred to a growth chamber at 22 °C (16/8 h photoperiod). 10-day-old in vitro-grown anti-hygromycin B seedlings were then transferred to soil mix. PCR and RT-PCR amplifications of the ClCASPL gene from candidate transgenic Arabidopsis plants were employed to confirm the successful transformation. WT and transgenic Arabidopsis T3 lines were used in this study. The SALK_034800C line was used for the screening of homozygous AtCASPL4C1 knockout mutants of Arabidopsis using the three primers sets designed from the online service (http://signal.salk.edu/tdnaprimers.2.html). Primers used in this study for cDNA amplification of ClCASPL gene and screening of AtCASPL4C1 knock-out mutant of Arabidopsis are listed in Supplementary Table 1.
Casparian Strip Analysis
Propidium iodide (PI) staining was used to check Casparian strip formation in root as previously published procedures9. Roots from 5-day-old seedlings grown in MS medium were incubated in the dark for 10 min in 15 μM (10 μg/ml) PI (Invitrogen) and then were rinsed twice in water. The stained roots were observed using a fluorescence microscope (OLYMPUS BX51, Japan). Excitation and emission wavelengths are 488 nm and 500–550 nm.
Chlorophyll Fluorescence Analysis
4-week-old seedlings of WT, AtCASPL4C1 and OX-ClCASPL of plants were cold treated at 10 °C and Fv/Fm measurements were taken. Individual plants were dark-adapted for 20 min prior to measurement. Chlorophyll fluorescence was measured in dark-adapted plants at 0, 6, 12, 24, 48 and 72 h under 10 °C cold stresses using a chlorophyll fluorescence system (M-Series Imaging-PAM, Germany).
Results
Characterization of CASPL
A CASP-like integral membrane protein gene, ClCASPL (Cla004012), was isolated in watermelon (Citrullus lanatus) using RT-PCR and genome blasting (Supplementary Figure 1-A). Bioinformatics analysis showed that ClCASPL is orthologous to At3g55390 from Arabidopsis (AtCASPL4C1, belonging to CASP family) (Fig. 1-A). Four transmembrane (TM) domains were predicted in ClCASPL (amino acids 45–67, 87–109, 130–149 and 169–191) and AtCASPL4C1 (At3g55390) proteins (amino acids 36–56, 78–98, 119–139 and 160–180 amino acid residues) by a variety of TM predication programs5 (Fig. 1-B). A total of 39 genes were identified that are defined as being part of the CASP family (UPF0497) in Arabidopsis, of which CASP1/2/3/4/5 has recently been identified to be associated with Casparian strip formation9. A phylogenetic tree for the CASP family in Arabidopsis21, defines 6 subfamilies using the Neighbor-Joining method and ClCASPL branches in the same subfamily as At3g55390 (AtCASPL4C1) of Arabidopsis (Fig. 1-C).
Characterization of the watermelon (Citrullus lanatus) ClCASPL and AtCASPL4C1 gene.
(A) Protein sequence alignment of ClCASPL with AtCASPL4C1. (B) The four transmembrane (TM) regions of ClCASPL and AtCASPL4C1 predicated by the TMHMM program. (C) Phylogenetic tree of ClCASPL with AtCASPL4C1 (At3G55390) and CASP family (UPF0497, 39 members, CASP1/2/3/4/5 have been identified as being involved with Casparian strip formation) from Arabidopsis.
Subcellular Localization of ClCASPL Protein
ClCASPL is predicted to be targeted to the plasma membrane, according to the TargetP and SignalP subcellular localization prediction programs22,23. The subcellular localization of ClCASPL was investigated by the generation of a ClCASPL-green fluorescent protein (GFP) fusion and transient expression in tobacco (Nicotianaben thamiana) leaves. Expression of the constructed ClCASPL-GFP gene revealed a fluorescence signal exclusively in the plasma membrane, co-localizing with the plasma membrane marker16 that was tagged with red fluorescent protein (RFP) (Fig. 2).
Expression Analysis of CASPL
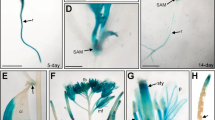
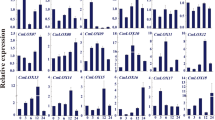
Transcriptomic analysis of public microarrays from the AtGenExpress developmental dataset5 and over seed germination17 indicated that AtCASPL4C1 (At3G55390) is expressed in various tissues, but unlike some genes in this family it could not be defined as being predominantly expressed in root tissues (Fig. 3, black box). In contrast, a strong root expression profile is observed for several genes in this family, including CASP1/2/3/4/5 and other CASP-like genes (Fig. 3, yellow box). To determine spatial expression more precisely AtCASPL4C1 expression patterns were determined using an AtCASPL4C1-promoter-GUS (for β-glucuronidase) reporter. Histochemical analysis expression in planta, with 12 independent lines all showed a very similar expression pattern, varied only slightly in the intensity of GUS staining. GUS-staining revealed that AtCASPL4C1 expression is visible in roots and leaves (Fig. 4A). We can observe AtCASPL4C1 expression in the vascular cylinder of roots, but no expression was detected in root tip (Fig. 4A,B). AtCASPL4C1 expression was also detected in emerged lateral root (Fig. 4C). The filament, stigma and sepal of flowers also displayed intense staining (Fig. 4D) and signal could also be readily detected in siliques, but not in seeds (Fig. 4E). These results indicate that AtCASPL4C1 is expressed in a wide range of variety of tissues and is not restricted to the vascular tissue of roots alone.
Transcript abundance profiles of 28 genes from the CASP protein family (UPF0497) that are represented on Affymetrix microarray chips.
Transcript abundance is shown over germination and development. Expression levels (log2) are shown for the 28 genes (represented on microarrays) that encoding CASP or CASP-like genes. Hierarchical clustering shows strong co-expression of specific CASP like genes with known CASP genes (strongly co-expressed genes indicated by an asterisk*). The transcript abundance for At3g55390 is shown in a black box and genes that display a root enriched expression profile are shown in a yellow box. White boxes indicate no expression was detected.
CASPL Negatively Regulates Growth Dynamics
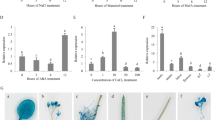
To investigate the function of CASPL in plants, AtCASPL4C1 knockout mutants and transgenic plants overexpressing ClCASPL in Arabidopsis were generated (Supplementary Figure 1 and 2). Growth and development parameters were measured to quantify any changes in phenotype due to the absence or over-expression of ClCASPL. The germination rate among wild type (Col-0), AtCASPL4C1 knockout and OX-ClCASPL plants did not significantly differ (Fig. 5A,C). The loss of AtCASPL4C1, however, did result in a small but significant increase in primary root length (Fig. 5D). In agreement with this, overexpressing ClCASPL in Arabidopsis significantly decreased primary root length (Fig. 5D). AtCASPL4C1 plants displayed significantly faster growth in several parameters, bigger plants (Fig. 5B,E), more biomass (Fig. 5H) and earlier rosette leaves development and flowering (Fig. 5F,G) compared to wild type (Col-0) and OX-ClCASPL plants. Thus, the loss of AtCASPL4C1 resulted in a positive growth affect, whereas over-expression gave the opposite effect. Thus, while only a single T-DNA insertion mutant is available for this gene in Arabidopsis, the complementary results of the knock-out and over-expressing plants strongly indicate that the phenotypes observed are due to the absence or increased abundance of CASPL.
Phenotypes of wild type, AtCASPL4C1 and OX-ClCASPL under normal growth condition.
(A) WT, AtCASPL4C1 and OX-ClCASPL growth for 5 days in MS medium. (B) Wild type, AtCASPL4C1 and OX-ClCASPL growth for 12 days. (C) Time course of germination for wild type, AtCASPL4C1 and OX-ClCASPL in medium. (D) Time course of primary root length for WT, AtCASPL4C1 and OX-ClCASPL in medium. (E) Wild type, AtCASPL4C1 and OX-ClCASPL growth for 28 days in Jiffy seedling culture substrate. (F) Wild type, AtCASPL4C1 and OX-ClCASPL growth for 42 days in Jiffy seedling culture substrate. (G) Time course of rosette leaves for wild type, AtCASPL4C1 and OX-ClCASPL in Jiffy seedling culture substrate. (H) Time course of dry weight for WT, AtCASPL4C1 and OX-ClCASPL in Jiffy seedling culture substrate. All growths are under growth condition of 22 °C and 16/8 h photoperiod in growth chamber. The star shows significance at 0.05 by Tukey test.
Casparian Strip Formation Analysis
In the CASP family (UPF0497) of Arabidopsis, CASP1/2/3/4/5 (Casparian strip membrane domain), were identified to mediate Casparian strip formation in the endodermis9. The Casparian strip is made up of a lignin polymer without suberin in Arabidopsis11. Detection of Casparian strip formation by lignin staining in the root endodermis of AtCASPL4C1, wild type and OX-ClCASPL clearly indicated lignin staining in root endodermis of AtCASPL4C1, WT and OX-ClCASPL (Supplementary Figure 3A). CASP1/2/3/4/5 transcript abundance was examined in root of wild type, AtCASPL4C1 and OX-ClCASPL. The transcript abundance of CASP1 was significantly altered, with increased transcript abundance in AtCASPL4C1 knock-outs and reduced abundance observed in OX-ClCASPL. The transcript abundance of CASP2, CASP3, CASP4 and CASP5 were significantly increased transcript abundance in AtCASPL4C1 knock-outs (Supplementary Figure 3B).
CASPL Negatively Mediated Plant Tolerance to Cold Stress
The transcript abundance of CASP protein family was examined under various kinds of stresses in Arabidopsis by transcriptomic analysis. These abiotic stresses result in significant up-regulation/down-regulation of the greatest number of CASP and CASP-like genes (Supplementary Figure 4).
The transcript abundance of ClCASPL and AtCASPL4C1 was examined under cold stress in watermelon and Arabidopsis. In watermelon, ClCASPL was induced and peaked in expression 12 h after cold treatment, while in Arabidopsis, AtCASPL4C1 was induced and peaked in expression 48 h after cold treatment (Fig. 6A). Moreover, the AtCASPL4C1 promoter-GUS activities after cold stress showed that expression of AtCASPL4C1 was induced by cold stress in Arabidopsis (Fig. 6B). To study the role of CASPL in cold tolerance, different parameters were used to evaluate cold tolerance in wild type, AtCASPL4C1 and OX-ClCASPL plants. Growth was monitored for 5-day-old plants exposed to cold at 10 °C for 7 days in medium. This examination showed that AtCASPL4C1 grows better under cold stress, based on the significantly longer primary root length (Fig. 7A–C). Furthermore, 3-week-old AtCASPL4C1 plants showed enhanced growth treated with cold stress, at 10 °C for 10 days (Fig. 7D,E), keeping relatively higher chlorophyll fluorescence parameters compared to WT (Fig. 7F), as well as, more rosette leaves and greater biomass (Fig. 7G,H). These results displayed greater cold tolerance than wild type and OX-ClCASPL.
CASPL gene expression after cold stress.
(A) Relative transcript abundance of ClCASPL in watermelon (real line) and AtCASPL4C1 in Arabidopsis (dash line) over the course of 72 h of cold stress at 10 °C. (B) GUS staining of CASPL-promoter after cold stress (10 °C) for up to 48 h in Arabidopsis seedlings.
Phenotypes of wild type, AtCASPL4C1 and OX-ClCASPL under cold stress.
(A) Wild type, AtCASPL4C1 and OX-ClCASPL seedling growth for 5 days at 22 °C in MS medium. (B) 5-day-old seedlings of wild type, AtCASPL4C1 and OX-ClCASPL grown for 7 days under cold stress at 10 °C in medium. (C) Time course of primary root length measurements for 5-day-old seedlings of wild type, AtCASPL4C1 and OX-ClCASPL grown for 7 days at 10 °C in medium. (D) Wild type, AtCASPL4C1 and OX-ClCASPL grown for 21 days at 22 °C in Jiffy seedling culture substrate. (E) 21-day-old seedlings of wild type, AtCASPL4C1 and OX-ClCASPL grown for 10 days at 10 °C in Jiffy seedling culture substrate. (F) Time course of Fv/Fm fluorescence values after cold treatment at 10 °C in 21-day-old seedlings of WT, AtCASPL4C1 and OX-ClCASPL plants. (G) Time course of rosette leaves for 21-day-old seedlings of WT, AtCASPL4C1 and OX-ClCASPL after cold stress at 10 °C in Jiffy seedling culture substrate. (H) Time course of dry weight for 21-day-old seedlings of WT, AtCASPL4C1 and OX-ClCASPL after cold stress at 10 °C in Jiffy seedling culture substrate. All growths are under growth condition of 16/8 h photoperiod in growth chamber. The star shows significance at 0.05 by Tukey test. OX represent OX-ClCASPL.
Discussion
The Casparian strip in the root endodermis is a barrier that restricts the free diffusion of molecules between the inner cell layers of the root. Recent studies revealed that this endodermal barrier is not only a regulator of water and nutrient uptake, but also probably acts as signal center for hormone-mediated control of growth24,25. It may be considered as a signal communication hub in response to the external environment, facilitating the activation of hormone signaling pathways and the propagation of calcium waves24,25. It also plays a central role in mediating cross talk between different cell layers during the development of new lateral organs, like lateral root emergence26. The molecular mechanisms involved in the deposition of the Casparian strip in endodermal cells are beginning to be uncovered27,28. The Casparian strip domain protein (CASP) anchors the plasma membrane to the cell wall and recruits the enzymes necessary for lignin biosynthesis27,28.
Evolutionary analysis indicated plant CASP family is associated with the MARVEL protein family described only in metazoans with similarity in specific transmembrane domains, in the overall tetraspan protein structure and putative Cys bridge in the second extracellular loop12. CASPs in this family are the earliest known proteins responsible for the CSD (Casparian strip membrane domain) formation9. The localizing at the CSD when expressed in the endodermis of CASPLs in this family prompts us to predict the role of yet-undiscovered CASPLs in mediating membrane subdomain formation and other plasma membrane domains in other cell types12. Here, we identified the membrane-localizing AtCASPL4C1 and ClCASPL genes that encode proteins of the CASP family. Overall, the absence of AtCASPL4C1 results in significantly increased growth under normal conditions. Based on analysis of CASP1/2/3/4/5 transcript abundance and lignin staining in roots, it is suggested that mutation in AtCASPL4C1 (At3G55390) did not dramatically affect the Casparian strip formation in roots. It has previously been shown in Atcasp1 or Atcasp3 single mutants, that Casparian strip formation was unaltered compared to WT, however, casp double mutants (casp1/casp3) displayed disorganized Casparian strip9. Thus, due to redundancy it is not possible to conclude if there is a direct role in Casparian strip formation for AtCASPL4C1. However, loss of function of AtCASPL4C1 was observed to result in earlier vegetative and reproductive development and increased biomass (Fig. 5).
Plants respond to environmental changes by altering tissue differentiation and development, referred to as plant phenotypic plasticity. Here, we showed CASPL gene also regulates enhanced growth under cold stress conditions. While AtCASPL4C1 belongs to this family based on predicted protein homology, the expression pattern based on in silico analyses and GUS reporter gene expression patterns suggest that the role of AtCASPL4C1 may extend well beyond the Casparian strip in roots (Figs 3 and 4). The widespread expression pattern in vascular tissues also suggests that it plays a more fundamental role in plant growth and development, which is supported by the altered phenotype when it is inactivated or over-expressed.
In conclusion, we identified a CASP-like ClCASPL gene in Citrullus lanatus and its ortholog AtCASPL4C1 in Arabidopsis are associated with growth and cold tolerance. These findings offered us clues to further explore the distinguished role of the Casparian strip protein family genes in growth, development and environmental communications.
Additional Information
How to cite this article: Yang, J. et al. A Casparian strip domain-like gene, CASPL, negatively alters growth and cold tolerance. Sci. Rep. 5, 14299; doi: 10.1038/srep14299 (2015).
References
Bohnert, H. J., Nelson, D. E. & Jensen, R. G. Adaptations to Environmental Stresses. The Plant Cell 7, 1099–1111 (1995).
Xiong, L. M., Schumaker, K. S. & Zhu, J. K. Cell signaling during cold, drought and salt stress. The Plant Cell 14, S165–S183 (2002).
Zhu, J. H., Dong, C. H. & Zhu, J. K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology 10, 290–295 (2007).
Mahajan, S. & Tuteja, N. Cold, salinity and drought stresses: an overview. Archives of Biochemistry and Biophysics 444, 139–158 (2005).
Schwacke, R. et al. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiology 131, 16–26 (2003).
Lucas, S., Dogan, E. & Budak, H. TMPIT1 from wild emmer wheat: first characterisation of a stress-inducible integral membrane protein. Gene 483, 22–28 (2011).
Garwe, D., Thomson, J. A. & Mundree, S. G. Molecular characterization of XVSAP1, a stress-responsive gene from the resurrection plant Xerophyta viscosa Baker. Journal of Experimental Botany 54, 191–201 (2003).
Jang, C. S. et al. The molecular characterization of a cDNA encoding the putative integral membrane protein, HvSec61 alpha, expressed during early stage of barley kernel development. Plant Science 168, 233–239 (2005).
Roppolo D. et al. A novel protein family mediates Casparian strip formation in the endodermis. Nature 473, 380–383 (2011).
Enstone, D. E., Peterson, C. A. & Ma, F. S. Root endodermis and exodermis: Structure, function and responses to the environment. Journal of Plant Growth Regulation 21, 335–351 (2002).
Naseer, S. et al. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proceedings of the National Academy of Science USA 109, 10101–10106 (2012).
Roppolo, D. et al. Functional and evolutionary analysis of the CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN family. Plant Physiology 165, 1709–1722 (2014).
Li, Y. M., Zhu, L., Xu, B. C., Yang, J. H. & Zhang, M. F. Identification of down-regulated genes modulated by an alternative oxidase pathway under cold stress conditions in watermelon plants. Plant Molecular Biology Reporter 30, 214–224 (2012).
Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469 (2003).
Sparkes, I. A., Runions, J., Kearns, A. & Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocal 1, 2019–2025 (2006).
Nelson, B. K., Cai, X. & Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant Journal 51, 1126–1136 (2007).
Narsai, R., Law, S. R., Carrie, C., Xu, L. & Whelan, J. In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiology 157, 1342–1362 (2011).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743 (1998).
Weigel, D. & Glazebook, J. Arabidopsis. A laboratory mannual. Cold Spring Harbor, NY: Cold Spring Harbor Press (2002).
Hong, J. K. & Hwang, B. K. The promoter of the pepper pathogen-induced membrane protein gene CaPIMP1 mediates environmental stress responses in plants. Planta 229, 249–259 (2009).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology Evolution 30, 2725–2729 (2013).
Emanuelsson, O., Nielsen, H., Brunak, S. & von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology 300, 1005–1016 (2000).
Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8, 785–786 (2011).
Robbins II, N. E., Trontin, C., Duan, L., Dinneny, J. R. Beyond the barrier: communication in the root through the endodermis. Plant Physiol. 166, 551–559 (2014).
Dinneny, J. R. A gateway with a guard: how the endodermis regulatd growth through hormone signaling. Plant Science 214, 14019 (2014).
Vermeer, J. E. M. et al. A spatial accommodation by neighboring cell is required for organ initiation in Arabidopsis. Science 343, 178 (2014).
Lee, Y., Rubio, M. C., Alassimone, J. & Geldner, N. A mechanism for localized lignin deposition in the endodermis. Cell 153, 402–412 (2013).
Hosmani, P. S. et al. Dirigent domian-containing proteins is part of the machinery requeired for formation of the lignin-based Casparian strip in the root. Proceedings of the National Academy of Science USA 110, 14498–14503 (2013).
Acknowledgements
This work was supported by the earmarked fund for Modern Agro-Industry Technology Research System of China (CARS-26-17), key science and technology breeding program of agricultural new variety of Zhejiang Province (2012C129031-2-11) and Science and Technology Program of Zhejiang province (2011C12001). We thank Dr. J Ni to offer the RFP markers.
Author information
Authors and Affiliations
Contributions
J.Y. and M.Z. conceived and designed the study. C.D., B.X., C.C., R.N. and H.Z. performed the experiments and analyzed the data. J.Y., J.W., M.Z. wrote the paper. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yang, J., Ding, C., Xu, B. et al. A Casparian strip domain-like gene, CASPL, negatively alters growth and cold tolerance. Sci Rep 5, 14299 (2015). https://doi.org/10.1038/srep14299
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep14299
This article is cited by
-
Genotyping-by-sequencing and genome-wide association study reveal genetic diversity and loci controlling agronomic traits in triticale
Theoretical and Applied Genetics (2022)
-
Global identification of long non-coding RNAs involved in the induction of spinach flowering
BMC Genomics (2021)
-
Silencing of StRIK in potato suggests a role in periderm related to RNA processing and stress
BMC Plant Biology (2021)
-
Molecular insights into the compatible and incompatible interactions between sugar beet and the beet cyst nematode
BMC Plant Biology (2020)
-
Root hairs enhance Arabidopsis seedling survival upon soil disruption
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.