Abstract
HIV-1 replication in macrophages can be regulated by cytokines and infection is restricted in macrophages activated by type I interferons and polarizing cytokines. Here, we observed that the expression levels of the cellular factors Trim5α, CypA, APOBEC3G, SAMHD-1, Trim22, tetherin and TREX-1 and the anti-HIV miRNAs miR-28, miR-150, miR-223 and miR-382 was upregulated by IFN-α and IFN-β in macrophages, which may account for the inhibiting effect on viral replication and the antiviral state of these cells. Expression of these factors was also increased by IFN-γ +/− TNF-α, albeit to a lesser extent; yet, HIV-1 replication in these cells was not restricted at the level of proviral synthesis, indicating that these cellular factors only partially contribute to the observed restriction. IL-4, IL-10 or IL-32 polarization did not affect the expression of cellular factors and miRNAs, suggesting only a limited role for these cellular factors in restricting HIV-1 replication in macrophages.
Similar content being viewed by others
Introduction
Macrophages are important target cells for HIV-1 and, due to their ubiquitous distribution and capacity to migrate into tissues, they contribute to the establishment of the viral reservoir1,2,3. Macrophages are long-lived cells that are resistant to the cytopathic effects of viral replication, allowing for continuous production and spread of viral particles for long periods of time4,5,6. Although the introduction of antiretroviral therapy has contributed to the control of viral replication in infected patients, the virus can persist in cellular reservoirs like macrophages, making eradication of the virus from the host thus far an impossible task7,8.
Macrophages become highly susceptible to HIV-1 infection after differentiation from circulating monocytes9,10,11. This gradual increase in HIV-1 susceptibility is not fully understood, but correlates with a decline in the expression of the anti-HIV cellular microRNAs (miR-28, miR-150, miR-223, miR-382) that target the HIV-1 genome12 and the innate restriction factors APOBEC3G and APOBEC3A13. After entering the tissue, macrophages encounter stimuli that shape their function. IFN-γ and TNF-α can skew macrophages into an M1 or pro-inflammatory phenotype. M2 or alternatively activated macrophages are induced by exposure to IL-4 or IL-13 (M2a) and IL-10 or glucocorticoids (M2c)14,15. HIV-1 replication in macrophages is modulated by cytokines that activate and/or polarize macrophages. In general, the effect of polarizing cytokines on HIV-1 replication in macrophages has been reported to be enhancing as well as inhibitory, depending on the state of the macrophages at the moment of infection. As an example, studies conducted previously showed that HIV-1 infection of IL-4 or IL-13 stimulated macrophages resulted in significantly reduced levels of reverse transcription and p24 production9,16,17. However, when infected macrophages were treated with IL-4, viral replication was enhanced9,18,19,20. Other studies have demonstrated inhibition of viral replication by stimulation of macrophages with IFN-γ, IL-4, IL-10 and the pro-inflammatory cytokine IL-3221,22,23,24,25,26,27,28. To date, the mechanism of HIV-1 inhibition in polarized macrophages remains unclear.
Recently, several factors involved in the intrinsic cellular defence against HIV-1 and other retroviruses have been described. These factors, including the tri-partite-containing motif (Trim) proteins Trim5α and Trim22, three prime repair exonuclease 1 (TREX1), SAM domain- and HD domain-containing protein 1 (SAMHD1), apolipoprotein B cytidine deaminase 3 (APOBEC3) G and tetherin29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44, interfere with HIV-1 replication at different steps in the viral life cycle. In this study, we investigated whether the inhibition of viral replication in cytokine stimulated MDM can be attributed to the expression of HIV-1 restriction factors, the cellular protein cyclophilin A (CypA) and microRNAs known to interfere with HIV-1 replication. We observed inhibition of HIV-1 infection in primary macrophages that were stimulated with IFN-α/β or polarized with IFN-γ +/− TNF-α, IL-4, IL-10 or IL-32, confirming previous studies. A comprehensive analysis of the expression of HIV-1-interfering factors in these cells revealed a strong induction by IFN-α and IFN-β, suggesting their involvement in controlling virus replication by type I interferons. In contrast, expression levels of these cellular factors and miRNAs increased to a lesser extend or remained unchanged in macrophages cultured with IFN-γ +/− TNF-α, IL-4, IL-10 or IL-32, suggesting a limited role for these factors in the observed restriction to HIV-1 replication in polarized MDM.
Results
Characterization of in vitro primary polarized monocyte-derived macrophages
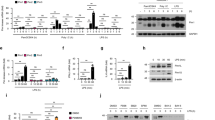
In vitro activation and polarization of MDM was evaluated by surface expression of CD14, CD16, CD64, CD80, CD163, CD200R (CD200 receptor) and CD206 (mannose receptor) on stimulated cells obtained from 4 different donors (Supplementary Figure S1). MDM cultured with type I interferons (IFN-α and IFN-β) showed high expression of CD80 and CD163. In agreement with previous reports14,45,46, MDM upregulated CD64 under M1 conditions (IFN-γ/TNF-α), CD200R and CD206 under M2a conditions (IL-4) and CD163 under M2c conditions (IL-10). Vice versa, we noticed downregulation of surface markers by specific cytokines, such as CD163 by IFN-γ/TNF-α and CD64 by IL-4. M-CSF had little impact on the analyzed cell surface markers, while MDM cultured with GM-CSF or IL-32 upregulated CD80 expression. Thus, by using a set of established cell surface markers, we could confirm phenotypic changes that had been evoked in the MDM by the indicated cytokines and growth factors.
HIV-1 replication in activated and polarized monocyte-derived macrophages
HIV-1 replication in MDM was analyzed by infecting MDM cultured for 5 days with differentiating factors (M-CSF, GM-CSF), type I interferons (IFN-α, IFN-β) or polarizing cytokines (IFN-γ +/− TNF-α, IL-4, IL-10, IL-32) with HIV-1 NL4-3 Ba-L. Viral replication was significantly inhibited in MDM activated with type I interferons as well as in IL-4- and IL-10-stimulated MDM, whereas virus replication was not affected in MDM stimulated with M-CSF, GM-CSF and IL-32 (Figure 1a). MDM stimulated with IFN-γ, either at a high concentration (250 U/ml) or at a low concentration (50 U/ml) in combination with TNF-α, also showed 3- to 4-fold reduction of HIV-1 replication, even though this was not statistically significant (Figure 1a).
HIV-1 replication in polarized MDM.
(a) MDM were treated with different cytokines and subsequently infected with NL4-3 Ba-L. Viral production was analyzed for 40 days after infection by p24 ELISA. The results are given as the mean fold induction of the cumulative p24 production, relative to the medium control (Student's t-test, *p<0.05, **p<0.01, ***p<0.001). The maximum p24 production in the medium control ranges from 277 to 689 ng/ml. Dots represent the values obtained from MDM isolated from different donors. (b) MDM treated with indicated cytokines were infected with the VSV-G pseudotyped NL4-3Luciferase virus and luciferase levels were measured 72 h post-infection. The results are given as the mean fold luciferase expression, relative to the medium control (Mixed linear model, repeated covariance: Diagonal, *p<0.05, **p<0.01, ***p<0.001). Dots represent the values obtained from MDM isolated from different donors.
To ensure efficient entry of HIV-1 in polarized MDM, parallel experiments were performed using a single round VSV-G pseudotyped luciferase reporter virus NL4-3Luc. A strong inhibition of infection was observed in MDM activated with type I interferon or polarized by IL-4, IL-10 or IFN-γ, either at a high concentration or at a low concentration in combination with TNF-α (Figure 1b). Infection with VSV-G/NL4-3Luc was also decreased in GM-CSF- and IL-32-stimulated MDM, contradicting our results using the replicating HIV-1 variant NL4-3 Ba-L, which suggests that the inhibitory effect of both GM-CSF and IL-32 might be submounted after prolonged culture.
Efficiency of HIV-1 reverse transcription in polarized and activated monocyte-derived macrophages
Next, we examined whether the process of reverse transcription occurred efficiently in polarized MDM. Stimulated cells were infected with NL4-3 Ba-L and analyzed by qPCR detecting early (R/U5) and relatively late (Pol) reverse transcription products at 48 h post infection (Figure 2). M-CSF, GM-CSF and IL-32 treatment had no effect on reverse transcription. A strong inhibition was observed in MDM treated with type I interferons, indicating that virus replication in these cells is inhibited at a very early step in the replication cycle (Figure 2a). IFN-γ polarization resulted in decreased levels of late products (Figure 2b), whereas treatment with a low concentration of IFN-γ in combination with TNF-α did not affect reverse transcription, indicating that inhibition of virus replication occurred at a later stage in the viral life cycle.
Efficiency of HIV-1 reverse transcription during infection of polarized MDM.
MDM were treated with the indicated cytokines and subsequently infected with NL4-3 Ba-L. Total DNA was isolated 48 h after infection. Early (a) and relative late (b) viral RT products were measured with qPCR detecting the R/U5 and pol region respectively. Levels of the RT products are expressed as the mean fold induction of the number of copies relative to the medium control, after normalization to β-actin for input (Student's t-test, *p<0.05, **p<0.01, ***p<0.001). Dots represent the values obtained from MDM isolated from different donors.
In IL-4-polarized MDM, inhibition of reverse transcription was demonstrated by a reduction of early and late reverse transcription products (Figure 2a, b). Efficient reverse transcription was observed in IL-10-polarized MDM, indicating that inhibition might occur at a later stage in the viral life cycle, probably at a post-translational level. These results also confirm previously described findings9,24.
Expression of cellular factors and anti-HIV miRNAs in polarized and activated monocyte-derived macrophages
Several cellular factors (TRIM5α, CypA, APOBEC3G, SAMHD1, TREX1, TRIM22 and tetherin) have been described to differentially affect viral infection at different levels in the replication cycle29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44. To determine if these cellular factors might be involved in the restricted replication of HIV-1 in polarized or activated macrophages, their expression levels were analysed at the moment of infection (5 days after isolation) by RT-qPCR (Figure 3). MDM stimulated with IFN-α or IFN-β expressed very high levels of all cellular factors analyzed (Figure 3). Increased expression levels of the cellular factors were also observed in MDM stimulated with IFN-γ with or without TNF-α, albeit to a lesser extent. In contrast, no increase in expression of any of these factors was observed in MDM maturated with M-CSF or GM-CSF, or polarized with IL-4, IL-10 or IL-32 (Figure 3). Certain culture conditions even led to a reduction in factor expression, such as M-CSF (Trim22), GM-CSF (TREX1 and tetherin), IL-4 (Trim22 and tetherin), IL-10 (Trim22) and IL-32 (CypA, TREX1 and tetherin).
Expression of HIV-1-interfering factors.
MDM were cultured in the presence of the indicated cytokines for 5 days. mRNA levels of CypA (a), Trim5α (b), APOBEC3G (c), TREX-1 (d), SAMHD-1 (e) and Trim22 (f) and tetherin (g) were measured with quantitative RT-PCR. Expression levels are given as the mean fold induction of the number of copies relative to the medium control, after normalization to β-actin for input (Student's t-test, *p<0.05, **p<0.01, ***p<0.001). Dots represent the values obtained from MDM isolated from different donors.
miRNAs have been recently described to target HIV-1 and regulate viral replication in monocytes and macrophages12. Here, we analyzed the expression of miR-28, miR-150, miR-223 and miR-382 by RT-qPCR in polarized and activated MDM at the moment of infection (Figure 4). In MDM activated with type I interferons, expression of all these miRNAs was upregulated (miR-150 and miR-382 up to 40 fold; miR-28 and miR-223 up to 6 fold). Furthermore, a slight increase in the expression of these miRNAs was observed in MDM treated with IFN-γ, with or without TNF-α. IL-4 and IL-10 polarization of MDM did not induce expression of any miRNA. IL-4 treatment even resulted in a downregulation of expression of these miRNAs. M-CSF, GM-CSF and IL-32 were also unable to induce expression of the miRNAs; moreover, a decreased expression of miR-150 was observed upon GM-CSF and IL-32 stimulation.
Expression of anti-HIV-1 miRNAs.
MDM were cultured in the presence of the indicated cytokines for 5 days. Expression levels of miRNA-28 (a) miRNA-150 (b), miRNA-223 (c) and miRNA-382 (d) were measured with quantitative RT-PCR and are expressed as the mean fold induction of miRNA copies relative to the medium control, after normalization to β-actin for input (Student's t-test, *p<0.05, **p<0.01, ***p<0.001). Dots represent the values obtained from MDM isolated from different donors.
Discussion
Macrophages play a crucial role in HIV-1 infection, especially in the establishment of the viral reservoir1,2,3. During HIV-1 infection, macrophages also contribute to the development of several tissue-specific diseases, including AIDS-related lymphomas, cardiovascular disease and HIV-1-associated neurocognitive disorders (HAND)47. Therefore it is of great importance to fully comprehend the involvement of macrophages in viral infection. We have assessed the effect of several cytokines on primary macrophages, in order to identify characteristics of the phenotypically different MDM, which may explain how these cells control HIV-1 replication. We have examined the effect of MDM stimulation by interferons and polarizing cytokines on the HIV-1 replication cycle, as well as on the expression of several cellular factors known to affect viral replication.
To achieve a better understanding of the effects of macrophage activation and polarization on the outcome of HIV-1 infection, in vitro, monocytes were allowed to differentiate into macrophages in the presence of growth factors, type I interferons and polarizing cytokines. Cell differentiation state was confirmed by differential expression of the membrane receptors CD14, CD16, CD64, CD80, CD163, CD200R and CD206, confirming previous observations45,46. We found that growth factors that mature monocytes into macrophages, such as M-CSF or GM-CSF, do not affect HIV-1 replication. In contrast, HIV-1 replication was strongly inhibited in MDM activated with IFN-α or β, or polarized with IFN-γ, with or without TNF-α, IL-4, IL-10 or IL-32, which is in agreement with previous studies9,16,17,22,23,24,25,26,27,28.
Infection of cytokine stimulated MDM with VSV-g-pseudotyped-HIV-1 single round virus was also strongly inhibited. The VSV-g envelope allows the virus to enter the cell via the endocytic pathway, instead of membrane fusion after binding of the HIV-1 envelop to CD4 and the CCR5 co-receptor, this indicates that the decreased HIV-1 infection in cytokine stimulated MDM is most likely not due to a restriction at the level of virus entry. Previously it was shown that endocytosis is differentially altered in polarized macrophages, i.e. IFN-γ and IL-10 reduce clathrin-mediated and mannose receptor-mediated endocytosis, whereas IL-4 is able to induce endocytosis48. However, IFN-γ and IL-10 stimulation did not reduce the level of reverse transcription products in MDM, indicating that these cytokines did not inhibit viral replication at the level of virus entry (Figure 2). Never the less, changes in the endocytic capacity of these cells, might explain some of the differences observed between the infection experiments performed with the wild type virus (Figure 1a) and the VSV-g pseudotyped single round reporter virus (Figure 1b).
Type I interferons blocked HIV-1 replication at the level of reverse transcription, confirming earlier studies49,50. We also observed that type I interferons were strong inducers of cellular factors that are able restrict HIV-1 replication (Trim5α, CypA, APOBEC3G, TREX1, SAMHD1, Trim22 and tetherin) and anti-HIV miRNAs (miR-28, miR-150, miR-223 and miR-382). Many of these factors are interferon-stimulated genes and have been reported to have synergistic effects in restricting viral infections51. It seems likely that the parallel induction of these cellular factors and anti-HIV miRNAs contributes to the observed strong inhibition of HIV-1 replication in type I interferon stimulated macrophages, especially those factors that are able to block the viral replication cycle before or at the process of reverse transcription (Trim5α, CypA, APOBEC3G, SAMHD1 and TREX1). Despite of their restrictive effect, HIV-1 is able to replicate in human cells in the presence of Trim5α and APOBEC3G and depends on certain levels of CypA for its replication. However, increased levels of these factors may contribute to the restricted infection observed in type I interferon treated macrophages. High concentrations of Trim5α and CypA can induce premature destabilization of the capsid proteins30,33,34,43, whereas high levels of APOBEC3G can inhibit accumulation of reverse transcription products35. APOBEC3 proteins levels are strongly enhanced by IFN-α and APOBEC3G is thought to be the most important inhibitory protein for HIV-1 produced by macrophages upon IFN-α stimulation, since its downregulation by siRNAs can rescue viral replication almost completely52. However, it has also been suggested that other Interferon stimulated genes (ISGs) can act together to inhibit early post entry steps of the viral replication of HIV-1 and other viruses49,51,52, supporting our hypothesis of a collective action of these restriction factors. Also, increased expression of SAMHD1 is likely to lower the deoxynucleotides levels and inhibit reverse transcription53, while TREX1 can degrade HIV-1 proviral DNA39. Trim22 and tetherin are most likely not crucial to the observed inhibition of HIV-1 replication elicited by type I interferons, because these cellular factors are known to interfere with later steps in the HIV-1 replication cycle (viral transcription and viral budding), whereas we observed an inhibitory effect of type I interferons at or prior to reverse transcription. The induction of restriction factors by IFN-β is less strong as compared to IFN-α. This might explain the high variation of viral infection observed with a single-round reporter virus.
It is remarkable that while the response elicited by type I interferons in macrophages is highly effective in stopping viral replication, these pathways are not activated in macrophages that encounter HIV-154. This has been attributed to TREX-1, which ensures successful viral replication by degrading the viral DNA and thus preventing its detection in the cytoplasm39. In dendritic cells, HIV-1 is restricted by SAMHD1 and interacts with CypA to prevent recognition by a cellular sensor, thus avoiding initiation of an innate response55. In contrast, SIV and HIV-2 are able to counteract the effect of SAMHD1 with Vpx, rescuing viral replication; however, this elicits interferon responses, which might explain the lower pathogenicity of these viruses in their natural host41. By losing Vpx, HIV-1 is able to escape from immune surveillance, since the low virus production in dendritic cells and macrophages will not elicit a strong innate response and as a result the virus gains an evolutionary advantage40,41,42.
When macrophages were polarized with IFN-γ, with or without TNF-α, we observed that reverse transcription occurred, indicating a restriction of viral replication at a later stage, compared to type I interferons. We also observed increased expression of all cellular factors analyzed, except for SAMHD1. However, the level of HIV-1 inhibition observed in these macrophages was highly variable but was not directly associated with the expression levels of the restriction factors. Thus, inhibition of HIV-1 in M1 macrophages is different from that induced by type I interferon and is perhaps mediated by other mechanisms, additional to restriction by the cellular factors investigated here.
In agreement with earlier observations9, HIV-1 replication in macrophages polarized with IL-4 towards an M2a phenotype was inhibited at the level of reverse transcription. In contrast, Cassol et al. observed that IL-4 stimulation of MDM results in a post-translational inhibition of HIV-1 replication22. This difference is probably due to the infection model of MDM that was used. We performed the infection on MDM differentiated for 5 days under cytokine stimulation, whereas in the study by Cassol et al., MDM were differentiated for 7 days and then stimulated with cytokines for 18 h, before infection. On the other hand, expression levels of the cellular factors affecting accumulation of HIV-1 reverse transcription products (CypA, Trim5α, APOBEC3G, TREX1 and SAMHD1) did not differ from those in unstimulated cells, indicating that none of these were involved in inhibiting replication of HIV-1 in IL-4-stimulated MDMs. Moreover, a decreased expression of Trim22, tetherin, miRNA-28, miRNA-150 and miRNA-223 was observed, which can possibly explain the increase in viral replication observed in other studies where IL-4 treatment was performed after HIV-1 infection9,18,19,20. Polarization into an M2c phenotype using IL-10, had no effect on HIV-1 reverse transcription and HIV-1 replication in these cells is most likely inhibited at a post-translational step24,26. Upon IL-10 treatment, a significant decrease of Trim22 expression was observed whereas the expression of the other cellular factors and miRNAs remained unchanged. Polarization with the proinflammatory cytokine IL-3256 also had no effect on the expression levels of the cellular factors and miRNAs analyzed here. Thus, the observed restriction of HIV-1 in M2a and M2c macrophages is not regulated by changes in the expression of the cellular factors and miRNAs analyzed here. Moreover, expression of other cellular proteins known to inhibit HIV-1 replication, such as APOBEC3A and p21 (CDKN1A)13,57, was also not associated with restriction of HIV-1 replication in M1, M2a or M2c macrophages (data not shown).
In summary, the strong inhibition of HIV-1 replication in type I interferon (IFN-α and IFN-β) activated macrophages is associated with induced expression of cellular proteins associated with restriction of HIV-1 replication (CypA, TRIM5α, APOBEC3G, SAMHD1, TREX1, TRIM22 and tetherin) and anti-HIV miRNAs. These factors combined probably contribute to the anti-HIV state of type I interferon-activated macrophages. However, these cellular factors and anti-HIV miRNAs are only partially or not responsible for the HIV-1 restriction in M1 and M2 macrophages. Our findings suggest that HIV-1 restriction in these cells may be mediated by a different yet unknown mechanism. Moreover, our data reveal the importance of macrophage activation and polarization on the outcome of HIV-1 infection. Further investigation of cellular mechanisms that interfere with HIV-1 replication in M1 and M2 macrophages is likely to identify new HIV-1 restriction factors and provide new insights into host-pathogen interactions.
Methods
Isolation of monocytes and cell culture
Monocytes were obtained from buffy coats from healthy blood donors. Written informed consents were obtained from all donors in accordance with the ethical principles set out in the declaration of Helsinki. This study was approved by the Medical Ethics Committee of the Academic Medical Center and the Ethics Advisory Body of the Sanquin Blood Supply Foundation in Amsterdam, The Netherlands. Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats using Lymphoprep (Axis-Shield, Olso, Norway) density gradient centrifugation. Monocytes were isolated by adherence to plastic and cultured in Iscove's modified Dulbecco's medium (IMDM; Lonza, Basel, Switzerland) supplemented with 10% [v/v] heat-inactivated human pooled serum (HPS), penicillin (100 U/ml; Invitrogen, Carlsbad, CA), streptomycin (100 µg/ml; Invitrogen) and ciproxin (5 μg/ml; Bayer, Leverkusen, Germany) for 5 days in the presence of different cytokines: IFN-α (250 U/ml; Sigma-Aldrich, St. Louis, MO, USA), IFN-β (250 U/ml; Sigma-Aldrich), IFN-γ (250 U/ml and 50 U/ml; Sigma-Aldrich) in combination with TNF-α (12.5 ng/ml; Peprotech, Rocky Hill, NJ, USA), IL-4 (50 ng/ml; Peprotech), IL-10 (50 ng/ml; Peprotech), GM-CSF (50 ng/ml; Peprotech), M-CSF (50 ng/ml; Sigma-Aldrich) or medium alone at 37°C in a humidified atmosphere supplemented with 5% CO2. At chosen concentrations, the cytokines showed a significant restriction of viral replication without affecting cell viability.
HEK293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) without Hepes (Lonza) supplemented with 10% [v/v] inactivated fetal calf serum, penicillin (100 U/ml) and streptomycin (100 μg/ml) and maintained in a humidified 10% CO2 incubator at 37°C.
Viruses and infection of monocyte-derived macrophages
HIV-1 NL4-3 Ba-L and single-round luciferase virus pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) were produced by transient transfection of HEK293T cells with pNL4-3 Ba-L or pNL4-3.Luc.R-E-construct in combination with pCMV-VSV-G using the calcium phosphate method58. Infectious virus was harvested at 48 and 72 h after transfection and filtered through a 0.22 μm filter. NL4-3 Ba-L titers were determined on PHA-stimulated PBMC as described previously59. Titers of the VSV-G pseudotyped single-round luciferase virus were determined on HEK293T cells.
For infection with HIV-1 NL4-3 Ba-L, MDM were cultured in the presence or absence of cytokines in 24-well plates (Nunc, Langenselbold, Germany) at a density of 500,000 cells per well and inoculated at day 5 with 10,000 TCID50 (equivalent to 13.5 ng p24 per 106 cells and an MOI of 0.02). After 24 h, the medium was changed to remove free virus particles. Supernatant was collected from the culture every 5–7 days for a period of 40 days post infection and analyzed for virus production by an in-house p24 capture ELISA60.
For infection with VSV-G pseudotyped NL4-3-luciferase reporter virus, MDM were cultured for 5 days in the presence or absence of cytokines in 96-well plates (Nunc) at a density of 50,000 cells per well and inoculated with 1,000 TCID50 (equivalent to an MOI of 0.02). At day 3 post infection, the supernatant was removed and 50 µl of luciferase substrate (0.83 mM ATP, 0.83 mM d-luciferin (Duchefa Biochemie B.V., Haarlem, The Netherlands), 18.7 mM MgCl2, 0.78 μM Na2H2P2O7, 38.9 mM Tris [pH 7.8], 0.39% [v/v] glycerol, 0.03% [v/v] Triton X-100 and 2.6 μM dithiothreitol) was added per well. Luminescence was measured for 1 sec per well, using a luminometer (Berthold Technologies, Bad Wildbad, Germany).
Quantitative PCR
Viral reverse transcription (RT) products were measured with real time quantitative PCR (qPCR). Total DNA was isolated from MDM inoculated with NL4-3 Ba-L 48 h post infection using L6 buffer (20 g of guanidine isothiocyanate in 100 ml 0.1 M Tris hydrochloride [pH 6.4], 22 ml 0.2 M EDTA [pH 8.0] and 2.5 g Triton X-100) as described previously61. Early and late RT products were measured using primers and probes detecting R/U5 and Pol products respectively61,62 and Roche Probes and Master mix (Roche, Basel, Switzerland). The following program was used in the LightCycler 480 Real-Time PCR System (Roche): denaturation: 95°C for 10 sec, amplification: 55 cycles of 95°C for 15 sec, 60°C for 60 sec. Serial dilution of the 8E5 cell line which contains 1 copy of HIV-1 proviral DNA per cell, was used as a standard curve for viral RT products. This cell line was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: 8E5/LAV from Dr. Thomas Folks63.
Expression levels of cellular factors and miRNAs were measured using RT-qPCR. RNA was isolated from 5 day MDM using TriPure Isolation Reagent (Roche). For mRNA quantification of cellular factors, cDNA was prepared using the M-MLV Reverse Transcriptase kit (Promega, Madison, WI) using an oligo dT primer. For reverse transcription and quantification of miRNAs, previously described stem-loop primers and qPCR primers for miR-28, miR-150, miR-223 and miR-382 were used64. The qPCR was performed with a Lightcycler 480 using specific primer pairs for cellular factors (Supplementary Table S1) and SYBR Green I Master (Roche). The following cycling conditions were used: denaturation: 95°C for 10 min; amplification: 50 cycles of 95°C for 10 sec, 58°C for 20 sec and 72°C for 30 sec. Purity of the PCR products was confirmed by melting curve analysis. β-actin expression levels were used to correct for cDNA input. A serial dilution of the 8E5 cell DNA was used as a standard curve for β-actin63.
Flow cytometry
MDM were fixed with 1% paraformaldehyde for 10 min at room temperature and subsequently washed with PBS containing 0.5% BSA, 10% 0.13 M trisodiumcitrate, 2 mM EDTA and 0.01% Na-azide. MDM were stained with the following antibodies for 30 min at 4°C in the dark: anti-human CD14-PerCP-Cy5.5 (eBioscience, San Diego, CA, USA), CD16-Alexa Fluor 647 (AbD Serotec, MorphoSys, Oxford, UK), CD64-Alexa Fluor 488 (Biolegend, San Diego, CA, USA), CD80-PE (BD Biosciences, San Jose, CA, USA), CD163-PE (BD Biosciences) CD200R-Alexa Fluor 647 (AbD Serotec), CD206-APC-Cy7 (Biolegend). Mouse IgG1κ conjugated to FITC, PE, PerCP-Cy5.5, APC and APC-Cy7 were used as isotype controls (BD Biosciences). Flow cytomery was performed with the FacsCanto II (BD Biosciences) and the results were analyzed in FlowJo, version 9.4.3 (Tree Star, Ashland, OR, USA).
Statistical analysis
Differences in p24 production, reverse transcription products and the expression levels of cellular factors or miRNAs were analyzed with a student t-test using GraphPad Prism 5 (GraphPad Software, La Jolla, California, USA). Luciferase values obtained from VSV-G/NL4-3-Luc infected MDM were analyzed using a mixed linear model (SPSS 16.0, Chicago, IL, USA). For the flow cytometry data, the geometrical means of each marker were calculated and compared between the different macrophage populations using a paired student's T-Test (GraphPad Prism 5).
References
Guadalupe, M. et al. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. J.Virol. 80, 8236–8247 (2006).
Poles, M. A. et al. Lack of decay of HIV-1 in gut-associated lymphoid tissue reservoirs in maximally suppressed individuals. J.Acquir.Immune.Defic.Syndr. 43, 65–68 (2006).
Chun, T. W. et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J.Infect.Dis. 197, 714–720 (2008).
Ho, D. D., Rota, T. R. & Hirsch, M. S. Infection of monocyte/macrophages by human T lymphotropic virus type III. J.Clin.Invest. 77, 1712–1715 (1986).
Nicholson, J. K., Cross, G. D., Callaway, C. S. & McDougal, J. S. In vitro infection of human monocytes with human T lymphotropic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV). J.Immunol. 137, 323–329 (1986).
Gendelman, H. E. et al. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J.Exp.Med. 167, 1428–1441 (1988).
Dornadula, G. et al. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 282, 1627–1632 (1999).
Fischer, M. et al. Residual HIV-RNA levels persist for up to 2.5 years in peripheral blood mononuclear cells of patients on potent antiretroviral therapy. AIDS Res.Hum.Retroviruses. 16, 1135–1140 (10-8-2000).
Schuitemaker, H. et al. Proliferation-dependent HIV-1 infection of monocytes occurs during differentiation into macrophages. J.Clin.Invest. 89, 1154–1160 (1992).
Rich, E. A., Chen, I. S., Zack, J. A., Leonard, M. L. & O'Brien, W. A. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1). J.Clin.Invest. 89, 176–183 (1992).
Sonza, S. et al. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J.Virol. 70, 3863–3869 (1996).
Wang, X. et al. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 113, 671–674 (2009).
Peng, G. et al. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 110, 393–400 (2007).
Mosser, D. M. & Edwards, J. P. Exploring the full spectrum of macrophage activation. Nat.Rev.Immunol. 8, 958–969 (2008).
Martinez, F. O., Helming, L. & Gordon, S. Alternative activation of macrophages: an immunologic functional perspective. Annu.Rev.Immunol. 27, 451–483 (2009).
Montaner, L. J., Bailer, R. T. & Gordon, S. IL-13 acts on macrophages to block the completion of reverse transcription, inhibit virus production and reduce virus infectivity. J.Leukoc.Biol. 62, 126–132 (1997).
Wang, J., Roderiquez, G., Oravecz, T. & Norcross, M. A. Cytokine regulation of human immunodeficiency virus type 1 entry and replication in human monocytes/macrophages through modulation of CCR5 expression. J.Virol. 72, 7642–7647 (1998).
Kazazi, F. et al. Recombinant interleukin-4 stimulates human immunodeficiency virus production by infected monocytes and macrophages. J.Gen.Virol. 73, 941–948 (1992).
Naif, H., Ho-Shon, M., Chang, J. & Cunningham, A. L. Molecular mechanisms of IL-4 effect on HIV expression in promonocytic cell lines and primary human monocytes. J.Leukoc.Biol. 56, 335–339 (1994).
Naif, H. M. et al. The state of maturation of monocytes into macrophages determines the effects of IL-4 and IL-13 on HIV replication. J.Immunol. 158, 501–511 (1-1-1997).
Kornbluth, R. S., Oh, P. S., Munis, J. R., Cleveland, P. H. & Richman, D. D. Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J.Exp.Med. 169, 1137–1151 (1989).
Cassol, E., Cassetta, L., Rizzi, C., Alfano, M. & Poli, G. M1 and M2a polarization of human monocyte-derived macrophages inhibits HIV-1 replication by distinct mechanisms. J.Immunol. 182, 6237–6246 (2009).
Cassol, E., Cassetta, L., Alfano, M. & Poli, G. Macrophage polarization and HIV-1 infection. J.Leukoc.Biol. 87, 599–608 (2010).
Kootstra, N. A., van't Wout, A. B., Huisman, J. G., Miedema, F. & Schuitemaker, H. Interference of IL-10 with HIV-1 replication in primary monocyte derived macrophages. J.Virol. 68, 6967–6975 (1994).
Montaner, L. J., Griffin, P. & Gordon, S. Interleukin 10 inhibits initial reverse transcription of human immunodeficiency virus type 1 and mediates a virostatic latent state in primary blood-derived human macrophages in vitro. J.Gen.Virol. 75, 3393–3400 (1994).
Naif, H. M., Chang, J., Ho-Shon, M., Li, S. & Cunningham, A. L. Inhibition of human immunodeficiency virus replication in differentiating monocytes by interleukin 10 occurs in parallel with inhibition of cellular RNA expression. AIDS Res.Hum.Retroviruses. 12, 1237–1245 (1996).
Nold, M. F. et al. Endogenous IL-32 controls cytokine and HIV-1 production. J.Immunol. 181, 557–565 (2008).
Rasool, S. T. et al. Increased level of IL-32 during human immunodeficiency virus infection suppresses HIV replication. Immunol.Lett. 117, 161–167 (2008).
Tissot, C. & Mechti, N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J.Biol.Chem. 270, 14891–14898 (1995).
Yin, L., Braaten, D. & Luban, J. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J.Virol. 72, 6430–6436 (1998).
Zhang, H. et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 424, 94–98 (2003).
Harris, R. S. et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 113, 803–809 (2003).
Sokolskaja, E., Sayah, D. M. & Luban, J. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J.Virol. 78, 12800–12808 (2004).
Stremlau, M. et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 427, 848–853 (26-2-2004).
Bishop, K. N., Holmes, R. K. & Malim, M. H. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J.Virol. 80, 8450-8458 (2006).
Bouazzaoui, A. et al. Stimulated trans-acting factor of 50 kDa (Staf50) inhibits HIV-1 replication in human monocyte-derived macrophages. Virology. 356, 79–94 (2006).
Barr, S. D., Smiley, J. R. & Bushman, F. D. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS.Pathog. 4, e1000007 (2008).
Neil, S. J., Zang, T. & Bieniasz, P. D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 451, 425–430 (2008).
Yan, N., Regalado-Magdos, A. D., Stiggelbout, B., Lee-Kirsch, M. A. & Lieberman, J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat.Immunol. 11, 1005-1013 (2010).
Goldstone, D. C. et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. (2011).
Laguette, N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 474, 654–657 (2011).
Hrecka, K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 474, 658–661 (2011).
Pertel, T. et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 472, 361–365 (2011).
Kajaste-Rudnitski, A. et al. TRIM22 inhibits HIV-1 transcription independently of its E3 ubiquitin ligase activity, Tat and NF-kappaB-responsive long terminal repeat elements. J.Virol. 85, 5183–5196 (2011).
Mantovani, A. et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25, 677–686 (2004).
Gordon, S. & Martinez, F. O. Alternative activation of macrophages: mechanism and functions. Immunity. 32, 593–604 (2010).
Deeks, S. G. HIV infection, inflammation, immunosenescence and aging. Annual Review of Medicine. 62, 141–155 (2011).
Montaner, L. J. et al. Type 1 and type 2 cytokine regulation of macrophage endocytosis: differential activation by IL-4/IL-13 as opposed to IFN-gamma or IL-10. J.Immunol. 162, 4606–4613 (15-4-1999).
Goujon, C. & Malim, M. H. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J.Virol. 84, 9254–9266 (2010).
Cheney, K. M. & McKnight, A. Interferon-alpha mediates restriction of human immunodeficiency virus type-1 replication in primary human macrophages at an early stage of replication. PLoS.One. 5, e13521 (2010).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 472, 481–485 (2011).
Peng, G., Lei, K. J., Jin, W., Greenwell-Wild, T. & Wahl, S. M. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J.Exp.Med. 203, 41–46 (23-1-2006).
Lahouassa, H. et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat.Immunol. 13, 223–228 (2012).
Tsang, J. et al. HIV-1 infection of macrophages is dependent on evasion of innate immune cellular activation. AIDS. 23, 2255–2263 (2009).
Manel, N. et al. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 467, 214–217 (2010).
Netea, M. G. et al. Interleukin-32 induces the differentiation of monocytes into macrophage-like cells. Proc.Natl.Acad.Sci.U.S.A. 105, 3515–3520 (2008).
Bergamaschi, A. et al. The CDK inhibitor p21Cip1/WAF1 is induced by FcgammaR activation and restricts the replication of human immunodeficiency virus type 1 and related primate lentiviruses in human macrophages. J.Virol. 83, 12253–12265 (2009).
He, J. et al. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J.Virol. 69, 6705–6711 (1995).
van't Wout, A. B., Schuitemaker, H. & Kootstra, N. A. Isolation and propagation of HIV-1 on peripheral blood mononuclear cells. Nat.Protoc. 3, 363–370 (2008).
Tersmette, M. et al. Detection and subtyping of HIV-1 isolates with a panel of characterized monoclonal antibodies to HIV-p24 gag. Virology. 171, 149–155 (1989).
Kootstra, N. A. & Schuitemaker, H. Phenotype of HIV-1 lacking a functional nuclear localization signal in matrix protein of gag and Vpr is comparable to wild-type HIV-1 in primary macrophages. Virology. 253, 170–180 (20-1-1999).
Butler, S. L., Hansen, M. S. & Bushman, F. D. A quantitative assay for HIV DNA integration in vivo. Nat Med. 7, 631–634 (2001).
Folks, T. M. et al. Biological and biochemical characterization of a cloned Leu-3- cell surviving infection with the acquired immune deficiency syndrome retrovirus. J.Exp.Med. 164, 280–290 (1986).
Huang, J. et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat.Med. 13, 1241–1247 (2007).
Acknowledgements
We thank John L. van Hamme and Martijn D.B. van de Garde for excellent technical assistance. This work has been supported by the Landsteiner Foundation for Blood Transfusion Research (grant 0435 and 0526) and the Academic Medical Center Amsterdam.
Author information
Authors and Affiliations
Contributions
V.C.J. designed and performed experiments, analyzed the data and wrote the manuscript. T.B. designed and performed experiments and contributed to writing of the manuscript. S.W.D.T. performed experiments and analyzed data. K.A.V.D. designed and performed experiments. M.A.N.R. designed and performed experiments. J.H. was involved in data analysis and writing of the manuscript. N.A.K. designed the study and the experiments and was involved in data analysis and writing of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Cobos Jiménez, V., Booiman, T., de Taeye, S. et al. Differential expression of HIV-1 interfering factors in monocyte-derived macrophages stimulated with polarizing cytokines or interferons. Sci Rep 2, 763 (2012). https://doi.org/10.1038/srep00763
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00763
This article is cited by
-
A Quantitative Approach to SIV Functional Latency in Brain Macrophages
Journal of Neuroimmune Pharmacology (2019)
-
Reduced antiretroviral drug efficacy and concentration in HIV-infected microglia contributes to viral persistence in brain
Retrovirology (2017)
-
NK cells control HIV‐1 infection of macrophages through soluble factors and cellular contacts in the human decidua
Retrovirology (2016)
-
Determinants in HIV-2 Env and tetherin required for functional interaction
Retrovirology (2015)
-
Severity of DSS-induced colitis is reduced in Ido1-deficient mice with down-regulation of TLR-MyD88-NF-kB transcriptional networks
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.