Abstract
The actinopterygians comprise nearly one-half of all extant vertebrate species and are very important for human well-being. However, the phylogenetic relationships among certain groups within the actinopterygians are still uncertain and debates about these relationships have continued for a long time. Along with the progress achieved in sequencing technologies, phylogenetic analyses based on multi-gene sequences, termed phylogenomic approaches, are becoming increasingly common and often result in well-resolved and highly supported phylogenetic hypotheses. Based on the transcriptome sequences generated in this study and the extensive expression data currently available from public databases, we obtained alignments of 274 orthologue groups for 26 scientifically and commercially important actinopterygians, representing 17 out of 44 orders within the class Actinopterygii. Using these alignments and probabilistic methods, we recovered relationships between basal actinopterygians and teleosts, among teleosts within protacanthopterygians and related lineages and also within acanthomorphs. These relationships were recovered with high confidence.
Similar content being viewed by others
Introduction
The actinopterygians (ray-finned fish) comprise approximately 28,000 extant species. This group is one of the major vertebrate groups, including nearly half of all extant vertebrate species1. Currently, according to molecular, morphological and paleontological studies, the actinopterygians, including 44 orders and 453 families1, are interpreted as a taxon comprising four major groups: cladistians, chondrosteans, holosteans and teleosteans2,3,4. Considerable effort has been made over a long time to resolve the phylogeny of actinopterygians based on both morphological and molecular data. However, the phylogenetic relationships among the major groups of actinopterygians were still controversial and unresolved, as are many of the proposed higher-level taxa within the Teleostei (e.g.,5,6). Debates on the ordinal relationships among basal euteleosts and on the most species-rich lineage, the Acanthomorpha, have long continued, although several new findings in molecular biology agree with results derived from morphological studies7,8,9. One of the major questions in actinopterygian phylogeny is the pattern of phylogenetic relationships among the higher “perch-like” fish, the order Perciformes and relatives (e.g.6,10,11). The monophyly of certain orders and families is in doubt and this difficulty creates even greater problems1.
Previous studies of actinopterygian phylogenies on the basis of nuclear genes focused primarily on particular groups and/or were usually based on relatively few markers. Even within the same species group, different gene markers have resulted in controversial phylogenies in certain cases. For example, MasonGamer and Kellogg found that gene trees of the grass tribe Triticeae resulting from four different single-gene data sets disagreed extensively in their intergeneric relationships12. Another study using four nuclear and two mitochondrial loci individually obtained different phylogenies among 17 Oriental Drosophila melanogaster species13. Rokas et al. selected 106 widely distributed orthologous genes from eight yeast genome sequences and concluded that a single or a small number of concatenated genes had a significant probability of supporting conflicting topologies, whereas more than 20 genes combined might yield a single, fully resolved species tree with maximum support14. Nevertheless, increasing the number of genes for accurate phylogenetic inferences inevitably constrains the number of analysed taxa and increases the percentage of missing data because of many limitations, such as time and resources. Furthermore, based on the aforementioned datasets used by Rokas et al., Phillips et al. obtained 100% supported but mutually incongruent trees using different tree-reconstruction methods and suggested that this inconsistency resulted from a compositional bias15. For all these reasons, phylogenomic approaches in systematics based on the analysis of multi-gene sequence data are becoming increasingly common because large numbers of characters and independent evidence from many genetic loci often result in well-resolved and highly supported phylogenetic hypotheses14,15,16. Furthermore, recent simulation and empirical studies have suggested that increases in gene sampling resulted in better performance than increases in taxon sampling17,18,19 and phylogenetic reconstruction appeared not to be sensitive to highly incomplete taxa as long as a sufficient number of characters were available20,21,22,23. Another advantage of phylogenomics is that the increasing throughput capacity of DNA sequencing technology has made available an ever-growing amount of sequence information, primarily in the form of large collections of expressed sequence tags (ESTs) or genome sequences. Phylogenetic inferences using a multi-locus approach, especially based on ESTs, are extensive because the use of ESTs can produce large numbers of gene sequences relatively easily and economically and can yield reliable and robust results24,25,26,27,28. Recently, Hittinger et al. sequenced transcriptomes of 10 mosquito species using the second-generation sequencing technologies and obtained robust phylogenetic inferences. They claimed this approach was an efficient, data-rich and economical option for generating large numbers of orthologous gene alignments for multi-locus phylogeny inference29. In view of these results, it is possible that robust phylogeny inferences for actinopterygians can be resolved by multi-gene approaches using multi-origin expression data.
Actinopterygians have been the group of vertebrates with the second best characterised genomes. Five fully sequenced and high-quality genomes are available for actinopterygians: Danio rerio (zebrafish), Gastroceus aculeatus (three-spined stickleback), Oryzias latipes (Japanese medaka), Takifugu rubripes (Japanese pufferfish) and Tetraodon nigroviridis (green spotted pufferfish). Additionally, many EST sequencing projects for a wide variety of teleost species have been conducted worldwide and hundreds of thousands of EST sequences are available. However, current deep phylogenetic studies of actinopterygians are primarily based on mitochondrial genomic data. Studies of this type based on nuclear genes are rare, especially in association with large-scale expression data. In the present study, the transcriptomes of three basal actinopterygians (Lepisosteus osseus, Polyodon spathula, and Polypterus delhezi) and two cypriniforms (Hypophthalmichthys molitrix, Hypophthalmichthys nobilis) were sequenced using the second-generation sequencing technologies (see Materials and Methods). Based on expression data generated in this study and on the results of previous genome and EST sequencing projects, we obtained multi-locus orthologous gene alignments for 17 of 44 orders within the class Actinopterygii. Subsequent analyses were performed to resolve the relationships among these species on the basis of these alignments.
Results
Sequence analyses and alignment
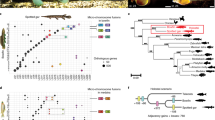
The transcriptome sequences used in this analysis for three basal actinopterygians and two cypriniforms were generated by us de novo (additional information in supplemental table S1). Transcriptome sequences, ESTs, mRNAs, Unigenes or cDNAs for 21 other species were downloaded from public databases (see methods). Based on these multi-origin expression data, we obtained 274 orhtologue groups (OGs) using OrthoSelect. The data profile for each species used in this study is shown in Table 1. Information for each OG (the number of species, length of alignment, percentage of missing data, best-fitting models of protein sequence evolution and accession number for each sequence) is given in supplemental table S2. The alignment files generated for phylogenetic analyses are given in supplemental file S1. The distribution of the alignment lengths of the 274 OGs is shown in Figure 1. The modal value of the alignment lengths appears to be in the range of 200–800 bp, with more than 90% shorter than 900 bp. Only 6 OGs had alignment lengths longer than 1000 bp and the mean length of all orthologues was 496 bp. There was a bias against obtaining longer alignments (the majority of the alignment lengths were approximately 500 bp). The reason for this outcome may be that most of our sequences were obtained directly from expression data rather than complete sequencing. The proportions of missing data for our OGs ranged from 10.0% for OG2806 to 62.7% for OG1174. The total number of OGs and percentages of missing data for each species are shown in Table 1. The missing data within these species ranged from 4.42% (Danio rerio) to 84.86% (Lepisosteus osseus). The nucleotide supermatrix concatenated from these 274 OGs included 135,969 bp and entirely missed 38.9% of the nucleotides. The average nucleotide composition of the concatenated supermatrix sequences was A = 27.1%, C = 24.6%, G = 27.0% and T = 21.3%.
Phylogeny inference based on nuclear multigenes
The concatenated nucleotide (excluding the third codon positions) and its conceptually translated amino acid genetic datasets were subjected to both Maximum Likelihood (ML, partitioned and unpartitioned) and Bayes Inference (BI, only unpartitioned) analyses and produced a consistent topology with similar phylogenetic support values. Almost all nodes were fully supported by posterior probabilities for BI. For ML, the node for the two perciforms, Dicentrarchus labrax (European seabass) and Sparus aurata (gilthead seabream), as sister group was not highly supported by the bootstrap values (Figure 2 and supplemental Figure S1 A–E). Both the AIC (Akaike information criterion) and the AICc values30 showed that the likelihood value with the partitioned supermatrix was better than the value with the unpartitioned supermatrix for the nucleotides. For the protein sequences, however, the likelihood value with the unpartitioned supermatrix was better than the value with the partitioned supermatrix. Interestingly, we reconstructed almost the same topology (supplemental SFigure 1 F and G) and the only difference was the placement of Oreochromis niloticus (Nile tilapia) based on the concatenated nucleotide supermatrix including the third codon positions. We recovered a monophyletic clade including Gasterosteus aculeatus (three-spined stickleback), Anoplopoma fimbria (sablefish), Sebastes caurinus (copper rockfish), Dissostichus mawsoni (Antarctic cod) and Hippoglossus hippoglossus (Atlantic halibut) with high confidence. Specifically, Gasterosteus aculeatus (Gasterosteiformes) and Anoplopoma fimbria (Scorpaeniformes) formed a sister-group relationship and Sebastes caurinus (Scorpaeniformes) and Dissostichus mawsoni (Perciformes) formed another monophyletic group with Hippoglossus hippoglossus (Pleuronectiformes) branched basal to this clade. The order Tetraodontiformes was placed as the most primitive taxon within Percomorpha (except Oreochromis niloticus). Figure 2 also shows that Fundulus heteroclitus and Oryzias latipes are sister, with Oreochromis niloticus branched basal to this clade. The monophyly and placement of major taxa such as Teleostei (Elopomorpha + Ostarioclupeomorpha or Otocephala + Euteleostei), Ostarioclupeomorpha (represented by Siluriformes + Cypriniformes), Acanthomorpha (Acanthopterygii (Atherinomorpha + Percomorpha) + Paracanthopterygii), which have been accepted extensively, were supported strongly by our analysis. The clade Protacanthopterygii ((Esociformes + Salmoniformes) + Osmeriformes) was recovered as monophyletic, with the Esociformes and the Salmoniformes as sister groups. As for the major actinopterygian clades, our results supported the topology (Polypteriformes, (Acipenseriformes, (Lepisosteiformes + Teleostei))).
The best-scoring maximum-likelihood (ML) tree derived from the concatenated supermatrix of the 274 nuclear genes (90,646bp, excluding the third codon positions) from the 26 actinopterygians with the GTRGAMMA model implemented in RAxML.
Numbers besides internal branches indicate bootstrap values based on 100 replicates. Other phylogenetic tree reconstruction strategies implemented in this report all obtained the same topology as this and are shown in supplemental Figure S1.
Discussion
The extant basal actinopterygians include four major lineages, the Polypteriformes, Acipenseriformes, Lepisosteiformes and Amiiformes. Although their basal positions within the actinopterygians have been consistently accepted by previous investigators1, considerable controversy over their relationship to the teleosts continues2,4,8. We conducted a comparative analysis of the phylogenetic positions of three lineages of basal actinopterygians (Polypteriformes, Acipenseriformes and Lepisosteiformes) relative to the teleosts with former hypotheses (please refer to Arratia 200131, who presented all possible morphological and molecular hypothesis and also to Arratia 200432). Our topology was in accordance with a previous conclusion based on gill-arch structure33 and with the first published significant hypothesis on the basal actinopterygian relationships based on molecular data34. Many recent conclusions based on morphological and molecular data were also consistent with our topology35,36,37. In contrast, previous findings that acipenseriforms or lepisosteiforms are more closely related to teleosts based on mitogenomic data8 or molecular synapomorphies38 were weakly supported by our topological test (Table 2). Currently, the polypteriforms (e.g., armored bichir) are widely accepted as the sister group of all other extant actinopterygians1. However, because the results we presented here did not include Amia calva in the analysis, this conclusion may be subject to bias and may require further investigation.
In addition to the basal actinopterygians, all other fishes in this study are collectively included within the Teleostei (Figure 2), which was represented by three main groups here: Elopomorpha, Ostarioclupeomorpha ( = Otocephala) and Euteleostei. Generally, researchers agreed that the protacanthopterygians occupy a phylogenetic position intermediate between the basal teleosts (ostarioclupeomorphs and below) and neoteleosts (stomiiforms and above)9 and are interpreted as basal Euteleostei. Because many of the morphological characters of the group have a mosaic distribution, the composition of this assemblage has undergone numerous changes over the past many decades1. Additionally, the deep relationships of the protacanthopterygians are so complex and controversial1,9 that at least 10 different phylogenetic hypotheses have been proposed (Figure 3 A–J; note that argentinoids are not shown because they are absent from our analysis. For more information, see Ishiguro's figure 1 A–J9, Springer & Johnson's figure 339 and Diogo's figure 240). Topological tests strongly suggested that our placement of the protacanthopterygians and related lineages was correct and confidently rejected other dichotomous ones (Table 2). Among these hypotheses, the phylogenetic position of the esociforms is one of the most controversial9,41. Our analysis strongly supports the hypothesis that the sister taxa of the esociforms were the salmoniforms rather than Neoteleostei39,42 or Osmeriformes40. This sister-group relationship is in accordance with many morphology-based and nearly all molecular-based hypotheses. Ramsden et al. corroborated this sister-group relationship from other perspectives, such as the life history and distribution of the fishes43. However, the placements of other lineages in these hypotheses are different from ours. For instance, the placement of Neoteleostei in our hypothesis is obviously different from the placement in earlier hypotheses except for that of Rosen44. Based on his morphological studies, Rosen suggested that protacanthopterygians were a monophyletic unit and that Protacanthopterygii and Neoteleostei formed a sister group (Fig 3A). This hypothesis is the same as ours. However, his placement of ostariophysans as a sister group to Protacanthopterygii and Neoteleostei was different from ours. Recently, several hypotheses based on mitochondrial data obtained the same topology as that found by our study. In fact, in the study of Ishiguro et al., the monophyly of protacanthopterygians cannot be rejected based on mitogenomic data if alepocephaloids are excluded and monophyly is enforced for the remaining groups of protacanthopterygians9. Before them, almost all morphology-based analyses consistently treated alepocephaloids and argentinoids, two suborders of the order Argentiniformes, as sister groups. However, Ishiguro et al.'s mitogenomic phylogenetic analysis argued that alepocephaloids were nested within the otocephalans with high statistical support9. Therefore, the phylogenetic position of these two lineages required further investigation.
Ten alternative phylogenetic hypotheses for basal euteleosts published after Rosen (1974).
A-H were modified from Ishiguro et al. (2003), I was modified from Diogo (2008) and J was modified from Fu (2010) and Broughton (2010). All terminal taxa were standardised to the three major protacanthopterygian lineages analysed in the present study (indicated by bold face).
Many taxa within the Euteleostei (minus Protacanthopterygii) that had true spines in the dorsal, anal and pelvic fins are included within the Acanthomorpha1. The superorder Acanthopterygii, which contains 13 orders, 267 families, 2,422 genera and approximately 15,000 species, can be divided into three large assemblages (termed Series, i.e., Mugilomorpha, Atherinomorpha and Percomorpha) and is the most species-rich superorder within this taxon1,45. Although many morphological and molecular studies have been conducted, the relationships among major lineages within the Acanthomorpha remain poorly defined1,6,7,10,11,45,46,47. In addition, certain orders and families within this assemblage are not monophyletic and this made the situation even worse1. In this study, we intended to test the possibility of recovering their relationships using many genes rather than resolving them thoroughly. The monophyly of the series Atherinomorpha, containing the Atheriniformes, Beloniformes (including the Adrianichthyoidei) and Cyprinodontiformes has been consistently suggested1,48. Similarly, Japanese medaka (Beloniformes) and killifish (Cyprinodontiformes) were grouped as sister groups with high confidence in this study. Moreover, we also recovered that one scorpaeniform fish was more closely related to the Antarctic cod (Perciformes), whereas the other scorpaeniform represented the sistergroup of three-spined stickleback (Gasterosteiformes). Certain species within Perciformes appeared more closely related to the orders Pleuronectiformes, Scorpaeniformes and Gasterosteiformes, but another species (Oreochromis niloticus) was more closely related to Atherinomorpha. This result is consistent with previous studies that proposed that Scorpaeniformes and Perciformes may not be monophyletic1,45,49. Interestingly, in a previous study based on mitogenomic sequences, Miya et al. found that internal branches among Percomorpha were only weakly supported but that members of Gasterosteiformes and Scorpaeniformes formed a strongly supported monophyletic group with a bootstrap value of 100%46. Moreover, the affinity of the cichlids with members of the Atherinomorpha has been consistently supported by studies based on nuclear genes17,50,51,52 and mitochondrial genomes35,37,48,53. This phylogenetic affinity is also supported by a unique egg morphology and spawning mode48. We recovered the tetraodontiforms as pre-perciforms with high confidence (Fig. 2). This result was in accordance with Springer and Johnson's finding, which was based on morphological studies39. However, evidence suggests that Scorpaeniformes (including the Dactylopteridae), Pleuronectiformes and Tetraodontiformes were most likely derivatives of perciform lineages1. Accordingly, our placement of Tetraodontiforms may be an artifact resulting from sparse taxonomic sampling of those species. Our multi-gene analysis recovered the relationships among most of these lineages. Nevertheless, many questions regarding the relationships among lineages within Acanthomorpha remain unanswered. For example, the monophyly of the Paracanthopterygii, the sister group of Atherinomorpha and Tetraodontiformes, the phylogenetic placement of Batrachoidiformes and the relationships among lineages within Percomorpha have long been controversial1. The last-named question poses particular difficulties because the monophyly of these groups is questionable and phylogenetic conclusions will depend on the choice of representatives50.
The deep phylogeny of actinopterygians is a long-standing and complex problem in the study of fish evolution. In this study, our taxon sampling for basal actinopterygians was purposefully chosen, but the information used for teleosts was based primarily on expression data available on public databases. We showed that phylogenomics based on integrating multi-origin expression data can recover their phylogeny with high confidence and that the major topology we obtained is consistent with that found by most previous studies. Moreover, the question of missing data is a significant problem for large-scale phylogenomic analysis. Philippe et al. showed that a supermatrix alignment with 25% missing data can still confidently resolve the phylogeny of eukaryotes21. In the case of actinopterygian phylogeny, an alignment with 38.9% missing data can result in a correct topology with high support. These results suggest that even with insufficient taxon sampling and several data gaps, large-scale phylogenomics based on integrating multi-origin expression data can produce a relatively good resolution of the the deep phylogeny of actinopterygians. Further investigations based on more purposefully chosen species may completely reconstruct the relationships of actinopterygians and provide a reliable phylogenetic framework for studying actinopterygian evolution.
Methods
Data collection and processing
Transcriptome sequences of five ray-finned fish species, Hypophthalmichthys molitrix (silver carp), Hypophthalmichthys nobilis (bighead carp), Lepisosteus osseus (longnose gar), Polyodon spathala (spoonbill cat) and the outgroup, Polypterus delhezi (armored bichir) were originally generated by Solexa sequencing in this study. Specimens of these species were purchased from a commercial source. The total RNA of each species was extracted from pooled organs with Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Poly (A+) RNA isolation, cDNA synthesis, preparation, sequencing (on an Illumina Genome Analyzer) and assembly (using the SOAP software package54) were performed at Beijing Genomics Institute. The assembled transcriptome sequences of European eel (Anguilla anguilla) were downloaded from EeelBase (http://compgen.bio.unipd.it/eeelbase/).
ESTs and/or mRNAs of Anoplopoma fimbria (sablefish), Dicentrarchus labrax (European seabass), Dissostichus mawsoni (Antarctic cod), Esox lucius (Northern pike), Hippoglossus hippoglossus (Atlantic halibut), Osmerus mordax (rainbow smelt), Sebastes caurinus (copper rockfish) and Sparus aurata (gilthead seabream), were downloaded from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov, GenBank status on 23 Dec 2009). Unigenes for Fundulus heteroclitus (killifish), Gadus morhua (Atlantic cod), Ictalurus furcatus (blue catfish), Ictalurus punctatus (channel catfish), Oreochromis niloticus (Nile tilapia), Pimephales promelas (fathead minnow) and Salmo salar (Atlantic salmon) were also downloaded from this database (GenBank status on 23 Dec 2009). Various contaminants and low-quality and low-complexity sequences within these data were screened and trimmed using SeqClean (http://compbio.dfci.harvard.edu/tgi/software) with NCBI's UniVec as a screening file.
Complementary DNA sequences of five model fish species, Danio rerio (zebrafish), Gasterosteus aculeatus (three-spined stickleback), Oryzias latipes (Japanese medaka), Takifugu rubripes (Japanese pufferfish) and Tetraodon nigroviridis (green spotted puffer), were retrieved from Ensembl (http://www.ensembl.org/, RELEASE62).
Sequence selection and alignment
Orthologue assignments were achieved using the slightly modified OrthoSelect method55 in this study. The default reference database of OrthoSelect was KOG (clusters of euKaryotic Orthologous Groups) and OrthoMCL, which included non-fish species. We know that teleosts have experienced the fish-specific genome duplication, which may result in “one2two” or “one2many” orthology relationships between teleosts and other species. To overcome this problem and to identify the orthology relationships unambiguously, we'd better use “one2one” orthology relationships as references. Therefore, we downloaded amino acid sequences of five model fish and their “one2one” relationships from Ensembl using BioMart. Each of these “one2one” sequence sets was termed an orthologue group (OG) in this study and the expression data were assigned to these OGs by a BLASTX analysis of individual EST sequences against all OG proteins. After the OG assignment, each sequence was translated using ESTScan56, GeneWise57 and a standard six-frame translation using BioPerl and aligned to the best hit from the previous BLAST search using bl2seq58. The translated sequence with the lowest E-value was chosen as the correctly translated sequence. Subsequently, one sequence from each organism was selected to represent the most probable ortholog to each other in accordance with their strategy based on matching positions normalized by its length in pairwise comparisons with MUSCLE59. However, because many ESTs were low-quality and included some frameshift errors or premature stopcodons, plus the limitations of bl2seq, we may discard the true ortholog in some species. To overcome these problems, we translated the expression data into protein sequences using ESTScan and found the best sequence from each database using hmmbuild and hmmsearch from the HMMER package60. After HMM selection, we obtained the orthology relationships for each OG. Then, we chose a model fish sequence and translated it into protein sequence and compared it to its orthologues separately with GeneWise (Only orthologue with a score more than 100 was retained). A customized Perl script was then used to extract matched nucleotides and to generate a sequence alignment for each OG. If a sequence was assigned to more than one OG, we discarded all these OGs to avoid any ambiguity. The OG alignments having more than 14 sequences were visually inspected and adjusted by hand using Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Finally, 274 OGs were selected and used for subsequent analyses.
Phylogenetic analysis
The nucleotides (excluding the third codon positions) and the conceptually translated amino acid alignments of these OGs were each concatenated, respectively. Both of the two supermatrices were subjected to subsequent Bayesian inference (BI) and Maximum Likelihood (ML) analyses. BI was performed with the MPI version of MrBayes 3.1.261, in which Markov Chain Monte Carlo (MCMC) calculations were spread across multiple CPUs and run on parallel computing architectures. The analysis was initiated from a random starting tree. Two runs with twelve chains of MCMC iterations were performed for 5 million generations (sampling trees every 100 generations) with the GTR + I + Γ models (for MrBayes and protein sequences, we used mixed+ I + Γ) of sequence evolution and the first 20,000 trees (2 million generations) were discarded as burn-ins. The average standard deviation of the split frequencies of the MCMC runs was used as the convergence diagnostic. The 50% majority-rule consensus tree was determined to calculate the posterior probabilities for each node. A parallel version of RAxML 7.2.662 was used for constructing Maximum Likelihood (ML) trees with the GTRGAMMA model for both the partitioned and the unpartitioned supermatrices (for the unpartitioned protein supermatrix, we used the PROTGAMMAJTTF model; the best fitting models of protein sequence evolution for each OG are listed in supplemental table S2). The partitioned supermatrices allow RaxML to assign different parameters for each gene. One hundred replicates for rapid bootstrap analyses62 were also performed with RAxML and a 50% majority rule consensus was calculated to determine the support values for each node. Fianlly, we placed the root at the branch quarter of Polypterus using MEGA563. The best-fitting models of protein sequence evolution were selected by ProtTest2.464. Tests of alternative phylogenetic hypotheses were implemented in CONSEL65.
References
Nelson, J. S. Fishes of the World, 4th Edition. Wiley, New York (2006).
Hurley, I. A. et al. A new time-scale for ray-finned fish evolution. Proceedings of the Royal Society B-Biological Sciences 274, 489–498 (2007).
Santini, F., Harmon, L. J., Carnevale, G. & Alfaro, M. E. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol Biol 9, 194 (2009).
Grande, L. An Empirical Synthetic Pattern Study of Gars (Lepisosteiformes) and Closely Related Species, Based Mostly on Skeletal Anatomy. The Resurrection of Holostei. Copeia, 1–863 (2010).
Lauder, G. V. & Liem, K. F. The evolution and interrelationships of the actinopterygian fishes. Bulletin of The Museum of Comparative Zoology 150, 103 (1983).
Miya, M. et al. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Molecular Phylogenetics and Evolution 26, 121–138 (2003).
Wiley, E. O., Johnson, G. D. & Dimmick, W. W. The interrelationships of Acanthomorph fishes: A total evidence approach using molecular and morphological data. Biochemical systematics and ecology 28, 319–350 (2000).
Inoue, J. G., Miya, M., Tsukamoto, K. & Nishida, M. Basal actinopterygian relationships: a mitogenomic perspective on the phylogeny of the "ancient fish". Molecular Phylogenetics and Evolution 26, 110–120 (2003).
Ishiguro, N. B., Miya, M. & Nishida, M. Basal euteleostean relationships: a mitogenomic perspective on the phylogenetic reality of the "Protacanthopterygii". Molecular Phylogenetics and Evolution 27, 476–488 (2003).
Chen, W. J., Bonillo, C. & Lecointre, G. Repeatability of clades as a criterion of reliability: a case study for molecular phylogeny of Acanthomorpha (Teleostei) with larger number of taxa. Molecular Phylogenetics and Evolution 26, 262–288 (2003).
Dettai, A. & Lecointre, G. Further support for the clades obtained by multiple molecular phylogenies in the acanthomorph bush. Comptes Rendus Biologies 328, 674–689 (2005).
MasonGamer, R. J. & Kellogg, E. A. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Systematic biology 45, 524–545 (1996).
Kopp, A. & True, J. R. Phylogeny of the oriental Drosophila melanogaster species group: A multilocus reconstruction. Systematic biology 51, 786–805 (2002).
Rokas, A., Williams, B. L., King, N. & Carroll, S. B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425, 798–804 (2003).
Phillips, M. J., Delsuc, F. & Penny, D. Genome-scale phylogeny and the detection of systematic biases. Molecular biology and evolution 21, 1455–1458 (2004).
Philippe, H., Lartillot, N. & Brinkmann, H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa and Protostomia. Molecular biology and evolution 22, 1246–1253 (2005).
Steinke, D., Salzburger, W. & Meyer, A. Novel relationships among ten fish model species revealed based on a phylogenomic analysis using ESTs. Journal of molecular evolution 62, 772–784 (2006).
Rokas, A. & Carroll, S. B. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Molecular biology and evolution 22, 1337–1344 (2005).
Rosenberg, M. S. & Kumar, S. Incomplete taxon sampling is not a problem for phylogenetic inference. Proc Natl Acad Sci U S A 98, 10751–10756 (2001).
Wolsan, M. & Sato, J. J. Effects of data incompleteness on the relative performance of parsimony and Bayesian approaches in a supermatrix phylogenetic reconstruction of Mustelidae and Procyonidae (Carnivora). Cladistics 26, 168–194 (2010).
Philippe, H. et al. Phylogenomics of eukaryotes: Impact of missing data on large alignments. Molecular biology and evolution 21, 1740–1752 (2004).
Driskell, A. C. et al. Prospects for building the tree of life from large sequence databases. Science 306, 1172–1174 (2004).
Wiens, J. J. Missing data, incomplete taxa and phylogenetic accuracy. Systematic Biology 52, 528–538 (2003).
Hughes, J. et al. Dense taxonomic EST sampling and its applications for molecular systematics of the Coleoptera (beetles). Molecular biology and evolution 23, 268–278 (2006).
Gerhold, D. & Caskey, C. T. It's the genes! EST access to human genome content. Bioessays 18, 973–981 (1996).
Witek, A. et al. EST based phylogenomics of Syndermata questions monophyly of Eurotatoria. BMC evolutionary biology 8, – (2008).
Roeding, F. et al. EST sequencing of Onychophora and phylogenomic analysis of Metazoa. Molecular phylogenetics and evolution 45, 942–951 (2007).
Dunn, C. W. et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–U745 (2008).
Hittinger, C. T., Johnston, M., Tossberg, J. T. & Rokas, A. Leveraging skewed transcript abundance by RNA-Seq to increase the genomic depth of the tree of life. Proceedings of the National Academy of Sciences of the United States of America 107, 1476–1481 (2010).
Posada, D. & Buckley, T. R. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53, 793–808 (2004).
Arratia, G. The Sister-group of Teleostei: Consensus and Disagreements. Journal of Vertertebrate Paleontology 21, 767–773 (2001).
ARRATIA, G. Mesozoic halecostomes and the early radiation of teleosts. In Arratia G, Tintori A (Eds.), Mesozoic Fishes 3 – Systematics, Paleoenvironments and Biodiversity. Verlag Dr. F. Pfeil, Muenchen, 279–315 (2004).
Nelson, G. J. Gill arches and the phylogeny of fishes : with notes on the classification of vertebrates. Bulletin of the American Museum of Natural History 141, 477–552 (1969).
Lê, H. L., Lecointre, G. & Perasso, R. A 28S rRNA-based phylogeny of the gnathostomes: first steps in the analysis of conflict and congruence with morphologically based cladograms. Mol Phylogenet Evol 2, 31–51 (1993).
Azuma, Y., Kumazawa, Y., Miya, M., Mabuchi, K. & Nishida, M. Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences. Bmc Evolutionary Biology 8 (2008).
Inoue, J. G., Kumazawa, Y., Miya, M. & Nishida, M. The historical biogeography of the freshwater knifefishes using mitogenomic approaches: A Mesozoic origin of the Asian notopterids (Actinopterygii: Osteoglossomorpha). Molecular Phylogenetics and Evolution 51, 486–499 (2009).
Setiamarga, D. H. E. et al. Divergence time of the two regional medaka populations in Japan as a new time scale for comparative genomics of vertebrates. Biology Letters 5, 812–816 (2009).
Venkatesh, B., Erdmann, M. V. & Brenner, S. Molecular synapomorphies resolve evolutionary relationships of extant jawed vertebrates. Proceedings of the National Academy of Sciences of the United States of America 98, 11382–11387 (2001).
Springer, V. G., Johnson G D. Study of the dorsal gill-arch musculature of teleostome fishes, with special reference to the Actinopterygii. Bulletin of the Biological Society of Washington 11, 200 (2004).
Diogo, R., Doadrio, I. & Vandewalle, P. Teleostean Phylogeny Based on Osteological and Myological Characters. International Journal of Morphology 26, 463–522 (2008).
López, J. A., Chen, W. J. & Orti, G. Esociform phylogeny. Copeia, 449–464 (2004).
Johnson, G. D. & Patterson, C. Relationships of lower euteleostean fishes. In: Stiassny M. L. J., Parenti, L. R., Johnson G. D. (Eds.), Interrelationships of Fishes. Academic Press, San Diego, CA, 251–332 (1996).
Ramsden, S. D., Brinkmann, H., Hawryshyn, C. W. & Taylor, J. S. Mitogenomics and the sister of Salmonidae. Trends in Ecology & Evolution 18, 607–610 (2003).
Rosen, D. E. Teleostean Interrelationships, Morphological Function and Evolutionary Inference. Amer. Zool. 22, 261–273 (1982).
Johnson, G. D. & Patterson, C. Percomorph Phylogeny - a Survey of Acanthomorphs and a New Proposal. Bulletin of Marine Science 52, 554–626 (1993).
Miya, M., Kawaguchi, A. & Nishida, M. Mitogenomic exploration of higher teleostean phylogenies: A case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Molecular Biology and Evolution 18, 1993–2009 (2001).
Miya, M., Satoh, T. R. & Nishida, M. The phylogenetic position of toadfishes (order Batrachoidiformes) in the higher ray-finned fish as inferred from partitioned Bayesian analysis of 102 whole mitochondrial genome sequences. Biological Journal of the Linnean Society 85, 289–306 (2005).
Setiamarga, D. H. et al. Interrelationships of Atherinomorpha (medakas, flyingfishes, killifishes, silversides and their relatives): The first evidence based on whole mitogenome sequences. Mol Phylogenet Evol 49, 598–605 (2008).
Smith, W. L. & Wheeler, W. C. Polyphyly of the mail-cheeked fishes (Teleostei : Scorpaeniformes): evidence from mitochondrial and nuclear sequence data. Molecular Phylogenetics and Evolution 32, 627–646 (2004).
Chen, W. J., Orti, G. & Meyer, A. Novel evolutionary relationship among four fish model systems. Trends in Genetics 20, 424–431 (2004).
Li, C. H., Lu, G. Q. & Orti, G. Optimal data partitioning and a test case for ray-finned fishes (Actinopterygii) based on ten nuclear loci. Systematic Biology 57, 519–539 (2008).
Li, B. et al. RNF213, a new nuclear marker for acanthomorph phylogeny. Molecular Phylogenetics and Evolution 50, 345–363 (2009).
Mabuchi, K., Miya, M., Azuma, Y. & Nishida, M. Independent evolution of the specialized pharyngeal jaw apparatus in cichlid and labrid fishes. Bmc Evolutionary Biology 7 (2007).
Li, R. Q., Li, Y. R., Kristiansen, K. & Wang, J. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008).
Schreiber, F., Pick, K., Erpenbeck, D., Worheide, G. & Morgenstern, B. OrthoSelect: a protocol for selecting orthologous groups in phylogenomics. BMC bioinformatics 10, – (2009).
Iseli, C., Jongeneel, C. V. & Bucher, P. ESTScan: a program for detecting, evaluating and reconstructing potential coding regions in EST sequences. Proceedings/... International Conference on Intelligent Systems for Molecular Biology; ISMB, 138–148 (1999).
Birney, E. & Durbin, R. Dynamite: a flexible code generating language for dynamic programming methods used in sequence comparison. Proc Int Conf Intell Syst Mol Biol 5, 56–64 (1997).
Tatusova, T. A. & Madden, T. L. BLAST 2 SEQUENCES, a new tool for comparing protein and nucleotide sequences. Fems Microbiology Letters 174, 247–250 (1999).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 (2004).
Durbin R, E. S., Krogh A. &. Mitchison G. Biological sequence analysis Cambridge, UK: Cambridge University Press. (2006).
Altekar, G., Dwarkadas, S., Huelsenbeck, J. P. & Ronquist, F. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20, 407–415 (2004).
Stamatakis, A., Hoover, P. & Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Systematic biology 57, 758–771 (2008).
Tamura, K. et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance and Maximum Parsimony Methods. Molecular Biology and Evolution 28, 2731–2739 (2011).
Abascal, F., Zardoya, R. & Posada, D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 (2005).
Shimodaira, H. & Hasegawa, M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247 (2001).
Acknowledgements
The authors were grateful to Henner Brinkmann for his critical reading of this manuscript and helpful comments and suggestions that greatly improved the paper. Thanks were also extended to the Wuhan sub-center of supercomputer environment, Chinese Academy of Science. This work was supported by a Grant from the Major State Basic Research Development Program of China (973 Program, no. 2007CB411601).
Author information
Authors and Affiliations
Contributions
M.Z. and S.P.H. conceived and designed the work. M.Z. and B.C.G. implemented the analysis and draft the manuscript. W.J.T. and G.A. participated in the analysis and revised the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplemental Table S1 and Supplemental Figure S1 and supplemental File S1
Supplementary Information
Supplemental table S2
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Zou, M., Guo, B., Tao, W. et al. Integrating multi-origin expression data improves the resolution of deep phylogeny of ray-finned fish (Actinopterygii). Sci Rep 2, 665 (2012). https://doi.org/10.1038/srep00665
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00665
This article is cited by
-
Phylogenomic Perspective on the Relationships and Evolutionary History of the Major Otocephalan Lineages
Scientific Reports (2018)
-
Evolution of Shh endoderm enhancers during morphological transition from ventral lungs to dorsal gas bladder
Nature Communications (2017)
-
Characterizing the transcriptome and molecular markers information for roach, Rutilus rutilus
Journal of Genetics (2016)
-
A comprehensive analysis of teleost MHC class I sequences
BMC Evolutionary Biology (2015)
-
RNA-Seq based phylogeny recapitulates previous phylogeny of the genus Flaveria (Asteraceae) with some modifications
BMC Evolutionary Biology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.