Abstract
Enteric pathogens can enter a persister state in which they survive exposure to antibiotics and physicochemical stresses. Subpopulations of such phenotypic dormant variants have been detected in vivo and in planta in the laboratory, but their formation in the natural environment remains largely unexplored. We applied a mathematical model predicting the switch rate to persister cell in the phyllosphere to identify weather-related stressors associated with E. coli and S. enterica persister formation on plants based on their population dynamics in published field studies from the USA and Spain. Model outputs accurately depicted the bi-phasic decay of bacterial population sizes measured in the lettuce and spinach phyllosphere in these studies. Predicted E. coli persister switch rate on leaves was positively and negatively correlated with solar radiation intensity and wind velocity, respectively. Likewise, predicted S. enterica persister switch rate correlated positively with solar radiation intensity; however, a negative correlation was observed with air temperature, relative humidity, and dew point, factors involved in water deposition onto the phylloplane. These findings suggest that specific environmental factors may enrich for dormant bacterial cells on plants. Our model quantifiably links persister cell subpopulations in the plant habitat with broader physical conditions, spanning processes at different granular scales.
Similar content being viewed by others
Introduction
Bacterial immigrants in the phyllosphere encounter heterogeneous physicochemical conditions dominated by solar radiation, water scarcity, temperature fluctuations, nutrient limitation, and antimicrobials that determine their fate in the plant environment [1,2,3,4]. Populations of human enteric pathogens inoculated onto crops in the field commonly undergo bi-phasic decay [5,6,7,8,9,10,11]. This trend is characterized by a large, rapid population decline after inoculation, followed by the survival of small subpopulations that remain relatively stable over time. In light of the recurrent outbreaks of enteric illness linked to fruit and vegetables [12,13,14], these few surviving cells are of high interest since they may represent subpopulations that serve as a reservoir for further amplification of the pathogen upon resumption of growth-permissive conditions on plants in the field or after harvest. Given the low infectious dose of pathogens such as Shiga toxin-producing E. coli (STEC) [15], initiation or resumption of pathogen multiplication on edible crops increases the probability of foodborne disease and the risk to public health.
The spatiotemporal heterogeneity of plant surfaces implies that clonal bacterial plant colonists may form subpopulations with distinct physiologies. Multiple bacterial pathways have been investigated at the single bacterial cell level on leaf surfaces and have been shown to vary greatly in activity among subpopulations of the inoculated reporter strain [1, 4, 16]. Given this heterogeneity, investigating the behavior of bacteria at the scale of single cells is critical to a comprehensive understanding of their ecology in the phytosphere. During bean leaf colonization by a Pantoea agglomerans bioreporter of cell replication, 3% of the viable cells did not divide once while the overall population of the bioreporter increased tenfold [17], suggesting the presence of a small subpopulation with distinct low metabolic activity. The role of phenotypic heterogeneity within clonal bacterial populations exposed to stressors has come to light through the discovery of persister subpopulations of human pathogens that survive antibiotic treatment without harboring mutations or genetic determinants imparting antibiotic resistance [18]. Mediated by metabolic dormancy likely resulting from ribosome inactivation [19], the persister state is viewed as a strategy to ensure the survival of a few in a population under stress. Current evidence supports that in addition to stochasticity, mechanistic processes induce persister cell formation in response to environmental cues via defined stress response pathways [20, 21]. Such cues include nutritional, acidic, oxidative, osmotic, and antimicrobials stresses. Importantly, although persister and viable-but-nonculturable (VBNC) cells may be part of a dormancy continuum, the persister state is transient and thus, cells may reverse from dormancy to a replicating state that allows for their recovery on culture media without the prolonged and specific resuscitation conditions required for culturing of VBNC cells [22].
We have recently reported the presence of E. coli O157:H7 (EcO157) persister cells after inoculation of the pathogen into irrigation water and spinach leaf wash water [23] and onto the lettuce phyllosphere of plants grown in the laboratory [24]. In the latter study, the pathogen population underwent bi-phasic decline on the leaves of lettuce plants exposed to dry conditions. Additionally, the persister cell fraction of the EcO157 population was greater under these dry conditions than during plant conditions that effected pathogen population growth or stability on the leaves. We further developed a mathematical model based on our laboratory persister data to describe EcO157 switch rates from nonpersister cells to persister cells on leaves of lettuce plants left to dry after inoculation. Applying our model equations from the decay regime on laboratory-inoculated plants, we estimated model parameters for four previously published field studies (in 2010–2013) of EcO157 survival on lettuce and obtained switch rates similar to those observed on lettuce in our laboratory study [24]. Hence, the model accurately described the survival rates of EcO157 in the lettuce phyllosphere in several field studies, all of which had shown bi-phasic survival of the pathogen [24].
The contamination of edible crops with enteric pathogens has become a major threat to public health and to the horticulture industry. Contaminated fruit and vegetables have caused numerous outbreaks of foodborne illness worldwide [12, 13, 25, 26]. In particular, leafy greens have been associated with large epidemics of STEC and S. enterica infections in the USA, Canada, and Europe [14, 27, 28]. Our understanding of the ecology of human enteric pathogens on plants in relation to environmental stressors dictating their behavior and physiology in planta in the field is essential to the development of strategies to enhance crop safety. In this study, we used mathematical modeling and data published by Belias et al. in 2020 on the survival of E. coli and S. enterica inoculated onto spinach and lettuce plants in field trials in California, New York, and Spain under known weather conditions [9] in order to 1) estimate the switch rate of the pathogens to persister cells on these crops in the field; and 2) explore the relationship between weather factors and the rate of persister cell formation in the phyllosphere.
Materials and methods
Case study
The experimental setup for the field studies that provided the bacterial population and weather data used here was previously described by Belias et al. [9]. Briefly, baby spinach and lettuce plants were spray-inoculated with E. coli and S. enterica (Salmonella) onto field plots established in Davis, CA (University of California, Plant Sciences Field Research Facility); Freeville, NY (Homer C. Thompson Research Farm, Cornell University); and Murcia, Spain (La Matanza Research Farm). The spinach and lettuce varieties were selected based on their suitability for baby leaf production: lettuce var. Tamarindo, and spinach var. Acadia F1 and Seaside F1. Four replicate trials at different times of the regional growing season were carried out per location. The plants were spray-inoculated with a 104 CFU/mL cocktail of rifampin-resistant strains of commensal E. coli and attenuated S. enterica serovar Typhimurium (Salmonella), and samples were collected for bacterial cell quantification by plate counts on selective and differential media at 0, 4, 8, 24, 48, 72 and 96 h post-inoculation. Concurrent with leaf sample collection, weather variables (temperature, relative humidity (RH), solar radiation intensity, and wind velocity) were recorded hourly for the respective field locations. The hourly dew point (DP) was calculated as a function of both the hourly temperature and RH.
Model for persister formation on plants
Mathematical modeling to characterize the switch rate from a non-persister bacterial cell (hereafter termed “normal cell”) to a persister cell in the phyllosphere under laboratory conditions was performed as described in our previously published study [24]. Briefly, persister cell fractions were quantified in culturable EcO157 populations after inoculation onto young lettuce plants cultivated in plant growth chambers. Persister cells recovered from the lettuce phyllosphere were identified using the antibiotic lysing method [23]. The greatest persister fraction in the EcO157 population on lettuce in our laboratory investigation above was observed during population decline on leaf surfaces of plants left to dry after inoculation. Using mathematical modeling, we calculated the switch rate from an EcO157 normal to persister cell on dry lettuce plants based on these data [24]. Importantly, our laboratory conditions mimicked inoculation conditions in which E. coli arrived via water on leaves, the surfaces of which progressively dried like under prevailing weather conditions in the field.
Based on the main dynamic observed in the field study data [9] and building on our previous study [24], we assumed that the total enteric pathogen population is composed of (i) non-persister (normal) cells consisting of two sub-populations, characterized by fast (n1) (CFU/100g) and slow (n2) (CFU/100g) decay, and (ii) the persister population, leading to the following model from Munther et al. [24]:
where \(\theta _{n_i}\)(1/h) is the death rate of the normal cells (subscript i = 1 for fast and i = 2 for slow), \(\hat p\) (CFU/100 g) represents the persister cell population at time t (h), \(\mu _{\hat p}\) (1/h) reflects the persister population inactivation rate, αd (1/h) is the switch rate from normal to persister state, βd (1/h) is the switch rate from persister to the normal state, and σ ∈ (0,1) is a constant, describing the fraction of persister cells switching back to the normal, slowly decaying state. Equation (1a) and (1b) reflect the assumption that times between switching states are exponentially distributed, using the expected values \(\frac{1}{{\alpha _d}}\) (h) and \(\frac{1}{{\beta _d}}\) (h) of the respective distributions.
Lacking data for potential persister populations from the field trials, we assumed the persister population is a fraction 1 > k > 0 of the tail population, as observed in Munther et al. [24]. Regarding the model above, this implies that \(\hat p \approx kn_2\) for \(t \ge t^ \ast\), where \(t^ \ast \approx \frac{1}{{\theta _{n_1}}}\) (the time scale of survival for the fast-decaying population (n1)). In accord with bi-phasic decay, for \(t \ge t^ \ast\), the main dynamics for slow decaying population (n2) is dictated by \(- \theta _{n_2}n_2\) in Eq. (1b). This suggests that the effective switch rates from n2 to \(\hat p\) and from \(\hat p\) back to n2 balance, so that \(\beta _d\sigma \hat p \approx \alpha _dn_2\) in Eq. (1b). Following these ideas, we simplified the model in Eq. (1a)–(1d) to:
where we ignored \(\beta _d\left( {1 - \sigma } \right)\hat p\) in (1a) since the decay rate (\(\theta _{n_1}\)) dominates. Also, by setting \(\theta _{\hat p} = \mu _{\hat p} + \beta _d(1 - \sigma )\), and using \(\beta _d\sigma \hat p \approx \alpha _dn_2\), we obtained Eq. (2c). Furthermore, because \(\hat p \approx kn_2\) for \(t \ge t^ \ast\), \(\theta _{\hat p} \approx\) \(\theta _{n_2}\).
In particular, the assumption that \(\hat p \approx kn_2\) for \(t \ge t^ \ast\) characterizes the switch rate from normal to persister cells, αd, as \(\alpha _d \approx k\alpha\), where α is a hypothetical switch rate assuming that the population is composed only of fast decaying normal cells (n1) and a hypothetical persister cell population (p). In this case, the hypothetical population p starts small at \(\widehat {p_0}\), initially increases due to switching from population n1 and then slowly decays as the n1 population is effectively inactivated (i.e., the tail of the total population is comprised entirely of p). From this perspective we utilized the following equations:
For mathematical justification regarding the relationship \(\alpha _d \approx k\alpha\), please see the appendix (Supplementary Information).
The utility of the relationship \(\alpha _d \approx k\alpha\), is twofold. First, we used model fitting (Eqs. (3a)–(3c)) to determine α from the respective field study data [9]. Note that using Eqs. (3a)–(3c), we actually fit for \(\theta _{n_1}\), θp, and α using the field study data [9]. Please reference the “model fitting procedure” section as well as the appendix for details concerning the unique determination of the aforementioned parameters, i.e., the practical identifiability of these parameters, and justification regarding the legitimacy of measured tail populations relative to the respective field trial data [9]. Second, because we wanted to examine Spearman’s correlations (corr) between αd and various weather factors, given a particular weather factor \(\vec w\) across trials \(i = 1, \ldots ,n\), let k be the maximum persister fraction (of the tail) across these n trials, that is, for each i, we have \(\alpha _{d_i} \approx k_i\alpha _i\), so \(\alpha _{d_i} \lesssim k\alpha _i\). Thus kαi represents the maximum persister switch rate for each trial i, and since corr(\(k\vec \alpha ,\vec w\)) =corr(\(\vec \alpha ,\vec w\)), we conducted the correlation analysis with the fitted α values in lieu of the actual persister switch rate αd.
The assumptions behind our approach are summarized below:
-
A.
The tails of pathogen populations surviving on plants in the field study [9] are comprised of some fraction k ∈(0,1) of persister cells since their decay rate is quite small and they remain culturable.
-
B.
Because \(\alpha _d \approx k\alpha\), we hereafter utilize α from model (3a)–(3c) as the representative persister switch rate.
-
C.
Given that the experimental context [24] for modeling persister switching occurred during population decline, we only employed trials from Belias et al. [9] that exhibited bi-phasic decay. Namely, we did not include trials in which significant bacterial growth was observed at the time scale of successive data points (the time scale in the field study is on the order of 4–16 h for the 1st day and then 24 h thereafter.)
-
D.
The switch rate from normal to persister cell is on average a monotonic function of some measure of environmental stress.
Based on assumptions A–D above, we applied the model (3a)–(3c) to published pathogen population size and weather data from four replicate trials in Spain, two in California, and one in NY [9]. More specifically, we fit model (3a)–(3c) to the respective population data in order to:
-
1.
determine values for the maximum switch rate α relative to the produce/bacteria type at the field scale,
-
2.
describe the correlative relationship between α and weather factors in the respective field trials.
Model fitting procedure
In model (3a)–(3c) above, we supposed dp/dtt = 0 > 0, i.e., we assumed that bacteria experience stress from the change in conditions from culture growth and inoculum suspension preparation to those on the plant surface and therefore, that persister formation increases in the phyllosphere immediately following inoculation. The report that EcO157 persister formation increases as early as 1 h after inoculation into leaf wash water [23], which could be considered as a proxy for the average oligotrophic environment that bacterial cells experience after spray inoculation onto leaves or through irrigation in the field, supports this assumption. To avoid identifiability issues between the initial persister population \(\widehat {p_0}\) and α regarding the model fits above, we assumed that \(\widehat {p_0}\)= 1 (\(\widehat {p_0}\) = 0 gives the same results). Thus, the initial persister population at inoculation is at its lowest, an assumption supported by Munther et al. [24], who observed an average fraction of EcO157 persisters of 0.0043% in the inoculum population. This imparts the largest possible switch rate, α, onto the population, corresponding to the largest and hence most conservative food safety risk.
Let yk (CFU/100 g of produce) be the average bacteria population measurement at time tk (h) and let Pk,X (CFU/100 g of produce) represent the model prediction (total population) at time tk relative to the parameter vector \(X = [ {\theta _{n_1} \,\ \theta_p \,\ \alpha } ]^T\). Following Eqs. (3a) and (3b), this means that \({{{{{{{\mathrm{P}}}}}}}}_{k,X} = n_1\left( {t_k,X} \right) + p(t_k,X)\). Since the population data spans multiple orders of magnitude, we calculated the residuals as \(e_{k,X} = \log _{10}y_k - \log _{10}P_{k,X}\). To determine the optimal model fit (see the appendix for details regarding a priori bounds on parameter ranges), we utilized the fminsearch function in MATLAB (MATLAB 2020b, The MathWorks, Inc., Natick, Massachusetts, United States) to determine the parameter vector X that minimizes the 2-norm of the following function F:
Correlation analysis
To provide a statistical foundation from which to relate the switch rate α and measured weather factors, we utilized Spearman and partial Spearman correlation. First, we calculated the Spearman correlation coefficients between α and each of the respective factors: 8-h average of temperature, RH, solar radiation, wind speed post-inoculation, and then we calculated the partial Spearman correlation coefficients for each respective weather factor, while controlling for the other three factors and simultaneously controlling for produce type (using lettuce =1 and spinach =0) (For details regarding why 8-h weather variables were used, see the “model fitting” subsection of the results.) The correlation coefficients were determined using the corr and partialcorr functions in MATLAB 2020b (The MathWorks, Inc., Natick, MA, USA). Considering the significant association of Salmonella α with RH and temperature, we also examined the correlation between α and dew point. Figure 1 presents a logical flow of the statistical analysis. Partial correlations with a P value of less than 0.05 were deemed significant. If the 8-h average of a weather factor exhibited a significant correlation with the switch rate, the 8-h minimum and range of the weather factor were also tested.
Factors in Step 1: UV (average ultraviolet radiation intensity), RH (average air relative humidity), Wind (average wind speed), and Temp (average air temperature). All weather data used in the statistical analysis were obtained over 8 h post-inoculation of E. coli and Salmonella onto lettuce and spinach leaves in the field.
Results and discussion
Model fitting
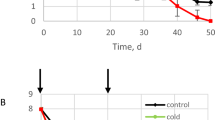
Model (3a)–(3c) describes the dynamics of bacterial persister formation after a population of cells arrive in an aqueous carrier (irrigation or inoculation solution) on leaf surfaces in the field and their physical and chemical environment subsequently changes due to gradually drying conditions in the phyllosphere. Figure 2 shows data obtained at all three locations, Spain, New York, and California in the field study by Belias et al. [9] and the model output resulting from fitting the model to these data. A rapid decline of E. coli and Salmonella population sizes was observed on both spinach and lettuce plants within 10–24 h post-inoculation. This early steep decline has been observed in several similar field studies about enteric pathogen survival on leafy vegetables [5,6,7, 10, 29, 30] as well as after inoculation of successful plant colonists such as Pseudomonas syringae and Pantoea agglomerans onto various plant species in the field [31, 32]. Whereas the bacterial epiphytes in the latter studies had the ability to multiply in the leaf habitat following this early decline, surviving enteric pathogens generally make up a small, and stable or slowly decaying population on the aerial surfaces of plants, similarly to trends shown in Fig. 2 and in other field studies cited above.
Sampling times were 0, 4, 8, 24, 48, 72, and 96 h post-inoculation. Each datum point represents the average population size and standard deviation from four subplots per trial from two, four, and one field trial(s) in California, Spain, and New York, respectively, as detailed in Materials and Methods. For details regarding the frequency of samples above the limit of detection (6 CFU/100g) for the tail populations refer to the “Details for parameter fitting” section in the appendix. The root mean square error (RMSE) from the optimal fits for population dynamics on each crop is provided as log10 CFU/100g in each figure.
Following procedures described above, we tested the fit of model (3a)–(3c) with the E. coli and Salmonella population dynamics observed in each of the field trials in California, New York and Spain (averaged across four field subplots per time point for each of the respective field trials, thus satisfying assumption C above). Figure 2 illustrates the average population sizes of each pathogen at each sampling time on lettuce and spinach leaves, demonstrating that the model captures the bi-phasic decay pattern. More specifically, the root mean square error from the optimal fits for E. coli dynamics on lettuce and spinach ranged from 0.0037 to 0.016 log10 CFU/100g and from 0.0039 to 0.013 log10 CFU/100g for Salmonella (Fig. 2) (Values of the parameters \(\theta _{n_1}\)(fast decay), θp (slow decay), and α (switch rate) from the fitting results illustrated in Fig. 2, are listed in Table S1 in the appendix.)
In order to obtain maximum information regarding the switch rate (α), we fit model (3a)–(3c) to field subplot data (rather than averaging across all subplots in the same field) for each respective trial at each location. From the results of the model fitting at the subplot level, \(T_{max}\) ≲ 10 h, that is, the persister population is predicted to reach a maximum within 10 h after the pathogen cells arrived on the leaves. Relative to this time scale, we only used weather information from the first 8 h post-inoculation in our analyses. From a mechanistic point of view this is illustrated in model Eq. (2c), as the switching mechanism (from normal to persister cells) dominates the persister population dynamics until the fast-decaying population becomes quite small, on a time scale proportional to \(1/\theta _{n_1}\). Notice that averaging across the fits from all trials, \(1/\theta _{n_1} \approx\) 16 h, which is relatively close to Tmax computed above.
E. coli α and weather parameters
Regarding direct correlations, the E. coli switch rate α was significantly correlated with the average 8-h temperature, RH, and solar radiation (Table 1). Partial rank correlation results (Table 1) indicated that only solar radiation and wind speed showed a significant correlation with the switch rate α while controlling for the other factors. Controlling for produce type in the partial correlation analyses did not have a substantial effect on the results. Thus, plant species was not a significant factor in the correlation between switch rate and stressor. This lack of difference between spinach and lettuce may be attributable to inoculation of the enteric pathogens onto the plants at an early growth stage (baby leaf) in the field studies by Belias et al. [9]. At this stage, both plant species would have young leaf tissue and an open canopy structure that may mediate overall similar physicochemical conditions experienced by bacterial cells on the phylloplane with respect to the weather stressors that we investigated. Indeed, Fig. 2 reveals similar enteric pathogen survival curves in both plants. It is probable that at maturity, plant species and in particular lettuce type would affect the results of a correlation analysis since the plant morphology varies greatly at that stage and this may generate differences in stresses from weather factors impacting bacterial cells on the leaf surface.
Table 1 indicates several important points. First, these results justify the importance of utilizing partial correlation to uncover interdependencies that may exist between multiple weather factors and the switch rate α. From this perspective, we placed more weight on the partial correlation results (rather than direct correlation), using these to provide statistical evidence in relation to established biological insight regarding the aforementioned assumption (D) and observed physical relationships between various weather factors. Contrasting results in Table 1, when controlling for the other respective weather factors, the correlations between the factors temperature and RH, and α did not remain statistically significant.
One possible explanation for temperature is its significant correlation with solar radiation and therefore temperature serves as a proxy for increased solar exposure. This conjecture is supported by the strength of the correlation between temperature and average solar radiation while controlling for RH (partial Spearman δ = 0.77, P = 9e-8). It is unclear why RH in the phyllosphere would not be correlated with the persister switch rate of E. coli. RH does not imply the presence or absence of free water on the phylloplane, since this would also depend on dew point; hence RH is not a direct measure of desiccation stress on bacterial cells. However, it is also plausible that desiccation is not an inducer of the persister state in E. coli since this has not been documented in the literature to date. Partial Spearman correlation was tested also between switch rate and the factors “temperature range”, “RH range”, and “dew point”, and results failed to show a significant association (data not shown). Therefore, contrary to what we describe below in Salmonella, none of the three tested weather factors that would contribute to the presence or absence of free water on the phylloplane showed any correlation with switch rate in E. coli in our study.
Solar radiation
Solar radiation showed a significantly moderate correlation with α, both directly and when controlling for the other weather factors and produce type. This positive correlation suggests that E. coli cells on lettuce and spinach leaves increasingly switch to the persister state with increasing solar radiation. The SOS response in E. coli, a ubiquitous bacterial strategy to repair DNA damaged by various stresses, including UV radiation, has previously been reported to induce persister cell formation [33]. As epiphytes, plant-associated bacteria benefit from DNA repair mechanisms such as those involving the SOS regulon, as well as pigmentation and spatial avoidance at particular microsites on the leaves, during colonization of leaf surfaces under solar radiation [34,35,36]. It was shown that protection of peanuts plants from UV-B lowered the relative abundance of UV-tolerant bacterial species in the phyllosphere community, suggesting that exposure to solar radiation enriched for UV-tolerant species [37]. Furthermore, Pseudomonas population sizes on baby spinach in the field correlated negatively with solar radiation providing further evidence that exposure to high radiation intensity imposes stress on bacterial cells [38]. Reduction of solar radiation exposure by shading plants [39] or inoculating the abaxial side of leaves [6] in the field improved E. coli O157:H7 survival in the spinach and lettuce phyllosphere, respectively; however, a direct correlation between population sizes and radiation intensity was not established. Overall, this suggests that UV exposure of bacterial cells in the phyllosphere may select for subpopulations composed at least partly of persister cells.
Wind velocity
The weak but significant negative association between wind velocity and α based on partial Spearman correlation in Table 1 is noteworthy since it implies that high wind speed inhibits the persister cell switch rate in E. coli on plant surfaces. High wind velocity, which disrupts the boundary layer of leaf surfaces [40] lowers the leaf temperature under conditions such as high solar radiation [41]. It may therefore alleviate stress on certain bacterial cells in the phylloplane and lower the formation rate of E. coli persisters. It is possible also that the movement of leaves in the canopy under high winds results in a shading effect and thus shorter exposure of the bacterial cells to UV radiation at any given time. The role of wind speed in the survival of bacterial residents on plants has not been investigated extensively. Average population sizes of Enterobacteriaceae and Pseudomonas spp. on spinach leaves in the field were reported to be negatively correlated with wind velocity [38]. Thus, while the population as a whole may be negatively impacted under high wind, a small subpopulation may experience a given stress alleviation mediated by this high wind velocity. One could hypothesize that the deleterious or beneficial effect of wind on bacteria in the phylloshere results indirectly from other environmental factors related to the disturbance of the laminar layer, where bacterial cells are located.
Salmonella α and weather parameters
The strongest correlations between α and measured weather factors were with RH, followed by solar radiation and temperature (Table 2). Comparing the results in Table 2 again illustrates the suitability of using partial Spearman correlation since direct correlation masks significant correlations between α and temperature and solar radiation.
Solar radiation
As observed also for E. coli switch rates (Table 1), solar radiation is positively correlated with the persister cell switch rate in Salmonella. Induction of persister cell formation by the SOS response has not been extensively investigated in Salmonella as in E. coli. However, the high similarity in their response network to DNA repair [42] would support a similar positive correlation of α in Salmonella with exposure to solar radiation in the phyllosphere.
RH and temperature
Contrary to our observations in E. coli, α is highly associated with RH in Salmonella. The negative correlation suggests that the persister formation rate decreases as RH in the lettuce and spinach environment increases. A relative humidity of 80% enhanced the survival of Salmonella, compared with 60%, in overall decaying populations on young lettuce plants in growth chamber studies [30], supporting this interpretation. Building on the significant results in Table 2, we calculated the partial correlations between α and minimum values and range for temperature, dew point, and RH, while controlling for the remaining three weather categories as well as produce type. Table 3 illustrates that each of these specific factors was significantly correlated with α. Minimum temperature and RH show a negative δ (partial Spearman correlation) value, likely due to the stress imposed onto Salmonella cells by cold temperature and dry air. In addition, the range in temperature, dew point, and RH over the 8-h period after inoculation used in our model was positively associated with the persister switch rate. Remarkably, it was also demonstrated by Lopez-Galvez et al. that large fluctuations in RH every 12 h negated the benefit of constant high RH in Salmonella survival on lettuce [30]. This observation and the results we describe herein support current thinking that large fluctuations in physicochemical conditions in the plant habitat impose additional stress onto bacterial residents [43]. It is unclear why the switch rate to persister cells in E. coli was not similarly affected by temperature and RH range. However, as neither the minimum nor the average values of these parameters were significantly associated with E. coli α based on partial Spearman correlation (Table 1), this may corroborate that temperature and RH were not significant triggers of the persister state in E. coli on plants in our study.
Whereas the direction of the correlation between α and solar radiation and RH makes intuitive sense since an increase in their magnitude would likely translate to an increase and decrease in cellular stress, respectively, the negative association of average temperature with Salmonella α (Table 2) requires more careful interpretation. The partial Spearman correlation between temperature and dewpoint (while controlling for RH) for Salmonella trials is very high (δ = 0.94, P value = 9.2e-16) (data not shown). Considering the monotonic relationship between temperature and dew point (given a fixed RH) [44], we can explain the negative correlation between temperature and α based on the strong negative correlation between dew point and α (Table 3). One could then surmise that increasing temperatures accompanied by leaf wetness support increasing multiplication of Salmonella in the phyllosphere, as demonstrated previously [45], and hence inhibit persister formation or cause a reversal from the persister to the normal state.
Dew point
Because temperature and RH are significantly associated with α, we further examined the correlation between α and dew point. Table 3 also lists partial correlation results between the various dew point factors and the switch rate α, while controlling for solar radiation, wind speed and produce type. Dew point average, minimum (results not shown) and range followed similar trends in correlation with α as those same parameters for RH.
The role of free water in bacterial cell behavior and viability on leaves cannot be overstated as it serves to ensure movement, multiplication, and acquisition of soluble nutrients, to mention only these few [46,47,48]. Lack of water may cause osmotic stress at microsites on leaves where solutes are at high concentrations [49]. It is noteworthy that we previously reported that population sizes of Salmonella declined and subsequently stabilized on dry leaves of cilantro plants maintained under low RH, then increased rapidly when dew formation was triggered on the leaves [50]. These dynamics may reflect the recovery of the small surviving subpopulation from the persister state. There is evidence that high osmolarity induces persister cell formation [21]. Furthermore, transient exposure to starvation enhances formation of persister cells in Salmonella [51] and an active starvation stress response mediates protection of Pseudomonas aeruginosa against several antibiotics [52]. Both osmotic stress and starvation may result from lack of free water, which concentrates solutes on leaves and restricts substrate availability to bacteria. Hence our model’s demonstration that Salmonella switch rate to persister cells on leaves is negatively correlated with dew point, which reflects the potential for the presence of free water on aerial plant surfaces, is consistent with current knowledge of persister formation in this pathogen.
Conclusions
The application of our persister model to E. coli and Salmonella survival data on plants from a previously published field study [9] and subsequent correlation analysis with weather parameters provide new knowledge of the physical factors that may enrich for bacterial persister cells in natural environments. Figure 3 illustrates the conclusions from our study. The strong positive association of predicted E. coli and Salmonella persister cell formation rates in the phyllosphere with solar radiation intensity accords with the reported induction of persistence by the SOS response in vitro in E. coli [33]. Likewise, the negative correlation of estimated Salmonella persister switch rates with weather parameters that promote water deposition on leaves is supported by previous observations that reversely, osmotic stress and starvation, both of which may be caused by low water availability in bacterial habitats, trigger the production of Salmonella persister cells [21, 51]. Our finding that the predicted switch rate of E. coli to the persister state decreased as wind velocity increased is worthy of further investigation since the role of wind speed in the bacterial ecology of plant surfaces remains unknown. Notably, the negative correlations of persister switch rate with wind and with weather factors related to water availability on surfaces were specific to E. coli and Salmonella, respectively, in this study. While the mechanisms involved in the formation of persisters must be elucidated further at the molecular level, there is evidence that they can differ among bacterial species. For example, Salmonella entered the persister state via specific pathways in response to inducers relevant to internalization into macrophages, while E. coli did not respond similarly [51]. Hence, our results corroborate that distinct pathways can be at play in persister cell formation in different organisms under various environmental pressures.
Since persister cells have enhanced survival to antimicrobial compounds, low pH, and reactive oxidative species [21], all of which underlie produce decontamination strategies, a better understanding of enteric pathogen physiology on crops allows for critical insight into the efficacy of different approaches to improve their safety for human consumption. New knowledge of the environmental factors that promote these subpopulations of dormant cells is not only of interest in public health but also more broadly informs microbial ecology. Our mechanistic model allowed for identification of weather parameters as an important measure of the predictable physiological state of a subpopulation of bacterial cells on plants. In a heterogeneous environment, such as the phyllosphere and other microbial habitats, the physiology of individual cells may determine the ultimate fate of the broader population over prolonged time scales. Bacterial persistence clearly adds to a spectrum of strategies that bacteria may employ to survive in the environment. These cells could therefore dictate the outcome of a range of environmental interventions, including microbiome engineering, bioremediation, protection against plant diseases with biocontrol agents, and antibiotic treatment, to mention only these few. Knowledge of the physicochemical factors that induce persister cells may serve as an important prediction tool. Thus, our study additionally proposes a new framework from which to investigate bacterial behavior in natural environments in the context of a problem that spans multiple scales and categories of information.
Data availability
All data used in this study can be found at https://github.com/abelias/die_off.
References
Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69:1875–83.
Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10:828–40.
Koskella B. The phyllosphere. Curr Biol. 2020;30:R1143–R6.
Meyer KM, Leveau JH. Microbiology of the phyllosphere: a playground for testing ecological concepts. Oecologia. 2012;168:621–9.
Bezanson G, Delaquis P, Bach S, McKellar R, Topp E, Gill A, et al. Comparative examination of Escherichia coli O157: H7 survival on romaine lettuce and in soil at two independent experimental sites. J Food Prot. 2012;75:480–7.
Erickson MC, Webb CC, Diaz-Perez JC, Phatak SC, Silvoy JJ, Davey L, et al. Surface and internalized Escherichia coli O157: H7 on field-grown spinach and lettuce treated with spray-contaminated irrigation water. J Food Prot. 2010;73:1023–9.
Moyne A-L, Sudarshana MR, Blessington T, Koike ST, Cahn MD, Harris LJ. Fate of Escherichia coli O157: H7 in field-inoculated lettuce. Food Microbiol. 2011;28:1417–25.
Williams TR, Moyne A-L, Harris LJ, Marco ML. Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS One. 2013;8:e68642.
Belias AM, Sbodio A, Truchado P, Weller D, Pinzon J, Skots M, et al. Effect of weather on the die-off of Escherichia coli and attenuated Salmonella enterica serovar Typhimurium on preharvest leafy greens following irrigation with contaminated water. Appl Environ Microbiol. 2020;86:e00899–20.
Gutiérrez-Rodríguez E, Gundersen A, Sbodio A, Koike S, Suslow TV. Evaluation of post-contamination survival and persistence of applied attenuated E. coli O157: H7 and naturally-contaminating E. coli O157: H7 on spinach under field conditions and following postharvest handling. Food Microbiol. 2019;77:173–84.
Lopez-Velasco G, Tomas-Callejas A, Sbodio AO, Pham X, Wei P, Diribsa D, et al. Factors affecting cell population density during enrichment and subsequent molecular detection of Salmonella enterica and Escherichia coli O157: H7 on lettuce contaminated during field production. Food Control. 2015;54:165–75.
Bennett SD, Sodha SV, Ayers TL, Lynch MF, Gould LH, Tauxe RV. Produce-associated foodborne disease outbreaks, USA, 1998–2013. Epidemiol Infect. 2018;146:1397–406.
Hackl E, Hölzl C, Konlechner C, Sessitsch A. Food of plant origin: production methods and microbiological hazards linked to food‐borne disease. Reference: CFT/EFSA/BIOHAZ/2012/01 Lot 1 (Food of plant origin with high water content such as fruits, vegetables, juices and herbs). EFSA Support Publ. 2013;10:402E.
Carstens CK, Salazar JK, Darkoh C. Multistate outbreaks of foodborne illness in the United States associated with fresh produce from 2010 to 2017. Front Microbiol. 2019;10:2667.
Paton JC, Paton AW. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11:450–79.
Remus‐Emsermann MN, Schlechter RO. Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytol. 2018;218:1327–33.
Remus-Emsermann MN, Leveau JH. Linking environmental heterogeneity and reproductive success at single-cell resolution. ISME J. 2010;4:215–22.
Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56.
Song S, Wood TK. ppGpp ribosome dimerization model for bacterial persister formation and resuscitation. Biochem Biophys Res Comm. 2020;523:281–6.
Wu Y, Vulić M, Keren I, Lewis K. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother. 2012;56:4922–6.
Gollan B, Grabe G, Michaux C, Helaine S. Bacterial persisters and infection: past, present, and progressing. Ann Rev Microbiol. 2019;73:359–85.
Ayrapetyan M, Williams TC, Oliver JD. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 2015;23:7–13.
Thao S, Brandl MT, Carter MQ. Enhanced formation of Shiga toxin-producing Escherichia coli persister variants in environments relevant to leafy greens production. Food Microbiol. 2019;84:103241.
Munther DS, Carter MQ, Aldric CV, Ivanek R, Brandl MT. Formation of Escherichia coli O157: H7 persister cells in the lettuce phyllosphere and application of differential equation models to predict their prevalence on lettuce plants in the field. Appl Environ Microbiol. 2020;86:e01602–19.
Lynch MF, Tauxe RV, Hedberg CW. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol Infect. 2009;137:307–15.
Coulombe G, Catford A, Martinez-Perez A, Buenaventura E. Outbreaks of Escherichia coli O157: H7 linked to romaine lettuce in Canada from 2008 to 2018: an analysis of food safety context. J Food Prot. 2020;83:1444–62.
Koutsoumanis K, Allende A, Alvarez‐Ordóñez A, Bover‐Cid S, Chemaly M, Davies R, et al. Pathogenicity assessment of Shiga toxin‐producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFSA J. 2020;18:e05967.
Marshall KE, Hexemer A, Seelman SL, Fatica MK, Blessington T, Hajmeer M, et al. Investigations of Shiga Toxin-producing Escherichia coli outbreaks linked to leafy greens, United States and Canada, 2009–2018. Emerg Infect Dis. 2020;26:2319–28.
Moyne AL, Harris LJ, Marco ML. Assessments of total and viable Escherichia coli O157: H7 on field and laboratory grown lettuce. PLoS One. 2013;8:e70643.
López-Gálvez F, Gil MI, Allende A. Impact of relative humidity, inoculum carrier and size, and native microbiota on Salmonella ser. Typhimurium survival in baby lettuce. Food Microbiol. 2018;70:155–61.
Beattie GA, Lindow SE. Comparison of the behavior of epiphytic fitness mutants of Pseudomonas syringae under controlled and field conditions. Appl Environ Microbiol. 1994;60:3799–808.
Sabaratnam S, Beattie GA. Differences between Pseudomonas syringae pv. syringae B728a and Pantoea agglomerans BRT98 in epiphytic and endophytic colonization of leaves. Appl Environ Microbiol. 2003;69:1220–8.
Dörr T, Lewis K, Vulić M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009;5:e1000760.
Jacobs JL, Carroll TL, Sundin GW. The role of pigmentation, ultraviolet radiation tolerance, and leaf colonization strategies in the epiphytic survival of phyllosphere bacteria. Microbial Ecol. 2005;49:104–13.
Gunasekera TS, Sundin GW. Role of nucleotide excision repair and photoreactivation in the solar UVB radiation survival of Pseudomonas syringae pv. syringae B728a. J Appl Microbiol. 2006;100:1073–83.
Sundin GW. Ultraviolet radiation on leaves: its influence on microbial communities and their adaptations. In: Lindow SE, Hecht-Poinar EI, Elliott VJ, editors. Microbiology of the phyllosphere. St-Paul, MN: APS Press; 2002. p. 27–41.
Jacobs JL, Sundin GW. Effect of solar UV-B Radiation on a phyllosphere bacterial community. Appl Environ Microbiol. 2001;67:5488–96.
Truchado P, Gil MI, Moreno-Candel M, Allende A. Impact of weather conditions, leaf age and irrigation water disinfection on the major epiphytic bacterial genera of baby spinach grown in an open field. Food Microbiol. 2019;78:46–52.
Wood JD, Bezanson GS, Gordon RJ, Jamieson R. Population dynamics of Escherichia coli inoculated by irrigation into the phyllosphere of spinach grown under commercial production conditions. Int J Food Microbiol. 2010;143:198–204.
Schreuder MD, Brewer CA, Heine C. Modelled influences of non-exchanging trichomes on leaf boundary layers and gas exchange. J Theor Biol. 2001;210:23–32.
Gates DM. Transpiration and leaf temperature. Ann Rev Plant Physiol. 1968;19:211–38.
Mérida-Floriano A, Rowe WP, Casadesús J. Genome-wide identification and expression analysis of SOS response genes in Salmonella enterica serovar Typhimurium. Cells. 2021;10:943.
Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev. 2000;64:624–53.
Lawrence MG. The relationship between relative humidity and the dewpoint temperature in moist air: a simple conversion and applications. Bull Am Meteorol Soc. 2005;86:225–34.
Brandl MT, Mandrell RE. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl Environ Microbiol. 2002;68:3614–21.
Doan HK, Ngassam VN, Gilmore SF, Tecon R, Parikh AN, Leveau JH. Topography-driven shape, spread, and retention of leaf surface water impacts microbial dispersion and activity in the phyllosphere. Phytobiomes J. 2020;4:268–80.
van der Wal A, Leveau JH. Modelling sugar diffusion across plant leaf cuticles: the effect of free water on substrate availability to phyllosphere bacteria. Environ Microbiol. 2011;13:792–7.
Beattie GA. Water relations in the interaction of foliar bacterial pathogens with plants. Annu Rev Phytopathol. 2011;49:533–55.
Axtell CA, Beattie GA. Construction and characterization of a proU-gfp transcriptional fusion that measures water availability in a microbial habitat. Appl Environ Microbiol. 2002;68:4604–12.
Brandl MT. Human pathogens and the health threat of the phyllosphere. In: Bailey JM, Lilley AK, Timms-Wilson TM, Spencer-Phillis PTN, editors. Microbial ecology of aerial plant surfaces. Wallingford, United Kingdom: CABI Publishing; 2006. p. 269–85.
Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–8.
Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–6.
Acknowledgements
The authors sincerely thank the anonymous reviewers for their careful reading and helpful suggestions for improving the exposition of the article. This study was supported by grant 2018CPS04 from the Center for Produce Safety and by the USDA Agricultural Research Service CRIS project 2030-42000-052-00-D.
Author information
Authors and Affiliations
Contributions
DSM and MTB conceived the paper. DSM and MTB drafted the original manuscript with RI. All authors contributed technical content and reviewed/edited versions of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brandl, M.T., Ivanek, R., Zekaj, N. et al. Weather stressors correlate with Escherichia coli and Salmonella enterica persister formation rates in the phyllosphere: a mathematical modeling study. ISME COMMUN. 2, 91 (2022). https://doi.org/10.1038/s43705-022-00170-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43705-022-00170-z