Abstract
As we age, structural changes contribute to progressive decline in organ function, which in the heart act through poorly characterized mechanisms. Taking advantage of the short lifespan and conserved cardiac proteome of the fruit fly, we found that cardiomyocytes exhibit progressive loss of Lamin C (mammalian Lamin A/C homolog) with age, coincident with decreasing nuclear size and increasing nuclear stiffness. Premature genetic reduction of Lamin C phenocopies aging’s effects on the nucleus and subsequently decreases heart contractility and sarcomere organization. Notably, Lamin C reduction downregulates myogenic transcription factors and cytoskeletal regulators, possibly via reduced chromatin accessibility. Subsequently, we find a role for cardiac transcription factors in regulating adult heart contractility and show that maintenance of Lamin C and cardiac transcription factor expression, prevents age-dependent cardiac decline. Our findings are conserved in aged nonhuman primates and mice, demonstrating that age-dependent nuclear remodeling is a major mechanism contributing to cardiac dysfunction.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Code availability
Software to image fly hearts, analyze their contraction and create kymographs (SOHA) is available at http://sohasoftware.com/index.html. Python code to assess lamin distribution is available at http://englea52.github.io/Englerlab/. Any ImageJ macros have been included in the supplementary software file.
References
Phillip, J. M., Aifuwa, I., Walston, J. & Wirtz, D. The mechanobiology of aging. Annu. Rev. Biomed. Eng. 17, 113–141 (2015).
Gilbert, H. T. J. & Swift, J. The consequences of ageing, progeroid syndromes and cellular senescence on mechanotransduction and the nucleus. Exp. Cell. Res. 378, 98–103 (2019).
Centers for Disease Control and Prevention. Underlying Cause of Death 1999–2020. https://wonder.cdc.gov/wonder/help/ucd.html (2022).
Sessions, A. O. et al. Extracellular matrix downregulation in the Drosophila heart preserves contractile function and improves lifespan. Matrix Biol. 62, 15–27 (2017).
Kaushik, G. et al. Vinculin network-mediated cytoskeletal remodeling regulates contractile function in the aging heart. Sci. Transl. Med. 7, 292ra99 (2015).
Sessions, A. O. & Engler, A. J. Mechanical regulation of cardiac aging in model systems. Circ. Res. 118, 1553–1562 (2016).
Birks, E. J. Molecular changes after left ventricular assist device support for heart failure. Circ. Res. 113, 777–791 (2013).
Van Berlo, J. H. et al. C-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 509, 337–341 (2014).
Cho, S., Irianto, J. & Discher, D. E. Mechanosensing by the nucleus: from pathways to scaling relationships. J. Cell Biol. 216, 305–315 (2017).
Janota, C. S., Calero-Cuenca, F. J. & Gomes, E. R. The role of the cell nucleus in mechanotransduction. Curr. Opin. Cell Biol. 63, 204–211 (2020).
Saucerman, J. J., Tan, P. M., Buchholz, K. S., McCulloch, A. D. & Omens, J. H. Mechanical regulation of gene expression in cardiac myocytes and fibroblasts. Nat. Rev. Cardiol. 16, 361–378 (2019).
Khatau, S. B. et al. A perinuclear actin cap regulates nuclear shape. Proc. Natl Acad. Sci. USA 106, 19017–19022 (2009).
Ramdas, N. M. & Shivashankar, G. V. Cytoskeletal control of nuclear morphology and chromatin organization. J. Mol. Biol. 427, 695–706 (2015).
Stephens, A. D., Banigan, E. J., Adam, S. A., Goldman, R. D. & Marko, J. F. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell. 28, 1984–1996 (2017).
Stephens, A. D. et al. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell. 29, 220–233 (2018).
van Steensel, B. & Belmont, A. S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017).
Dahl, K. N., Ribeiro, A. J. & Lammerding, J. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102, 1307–1318 (2008).
Chatzifrangkeskou, M., Kah, D., Lange, J. R., Goldmann, W. H. & Muchir, A. Mutated lamin A modulates stiffness in muscle cells. Biochem. Biophys. Res. Commun. https://doi.org/10.1016/j.bbrc.2020.05.102 (2020).
Verstraeten, V. L. R. M., Ji, J. Y., Cummings, K. S., Lee, R. T. & Lammerding, J. Increased mechanosensitivity and nuclear stiffness in Hutchinson–Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell 7, 383–393 (2008).
Lammerding, J. et al. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 281, 25768–25780 (2006).
Srivastava, L.K., Ju, Z., Ghagre, A. & Ehrlicher, A. J. Spatial distribution of lamin A/C determines nuclear stiffness and stress-mediated deformation. J. Cell Sci. 134, jcs248559 (2021).
Buxboim, A. et al. Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Curr. Biol. 24, 1909–1917 (2014).
Dahl, K. N. et al. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. USA 103, 10271–10276 (2006).
Brandt, A. et al. Developmental control of nuclear size and shape by kugelkern and kurzkern. Curr. Biol. 16, 543–552 (2006).
Jevtić, P. et al. Concentration-dependent effects of nuclear lamins on nuclear size in Xenopus and mammalian cells. J. Biol. Chem. 290, 27557–27571 (2015).
Pajerowski, J. D., Dahl, K. N., Zhong, F. L., Sammak, P. J. & Discher, D. E. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl Acad. Sci. USA 104, 15619–15624 (2007).
Bonne, G. et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 21, 285–288 (1999).
Di Barletta, M. R. et al. Different mutations in the LMNA gene cause autosomal dominant autosomal recessive Emery-Dreifuss muscular dystrophy. Am. J. Hum. Genet. 66, 1407–1412 (2000).
Taylor, M. R. G. et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J. Am. Coll. Cardiol. 41, 771–780 (2003).
Capell, B. C., Collins, F. S. & Nabel, E. G. Mechanisms of cardiovascular disease in accelerated aging syndromes. Circ. Res. 101, 13–26 (2007).
Scaffidi, P. & Misteli, T. Reversal of the cellular phenotype in the premature aging disease Hutchinson–Gilford progeria syndrome. Nat. Med. 11, 440–445 (2005).
Liu, B. et al. Genomic instability in laminopathy-based premature aging. Nat. Med. 11, 780–785 (2005).
Goldman, R. D. et al. Accumulation of mutant lamin A progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. USA 101, 8963–8968 (2004).
Shumaker, D. K. et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl Acad. Sci. USA 103, 8703 (2006).
Haithcock, E. et al. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 102, 16690–16695 (2005).
Brandt, A., Krohne, G. & Großhans, J. The farnesylated nuclear proteins kugelkern and lamin B promote aging-like phenotypes in Drosophila flies. Aging Cell 7, 541–551 (2008).
Scaffidi, P. & Misteli, T. Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063 (2006).
Larson, K. et al. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. https://doi.org/10.1371/journal.pgen.1002473 (2012).
McClintock, D. et al. The mutant form of lamin a that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS ONE 2, e1269 (2007).
Messner, M. et al. Upregulation of the aging related LMNA splice variant progerin in dilated cardiomyopathy. PLoS ONE 13, e0196739 (2018).
Afilalo, J. et al. Age-related changes in lamin A/C expression in cardiomyocytes. Am. J. Physiol. Circ. Physiol. 293, H1451–H1456 (2007).
Freund, A., Laberge, R. M., Demaria, M. & Campisi, J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 23, 2066–2075 (2012).
Chen, H., Zheng, X. & Zheng, Y. Age-associated loss of lamin-B leads to systemic inflammation and gut hyperplasia. Cell 159, 829–843 (2014).
Han, L. et al. Lamin B2 levels regulate polyploidization of cardiomyocyte nuclei and myocardial regeneration. Dev. Cell https://doi.org/10.1016/j.devcel.2020.01.030 (2020).
Nikolova, V. et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J. Clin. Invest. 113, 357–369 (2004).
Cammarato, A. et al. A mighty small heart: the cardiac proteome of adult Drosophila melanogaster. PLoS ONE 6, e18497 (2011).
Nishimura, M. et al. A dual role for integrin-linked kinase and β1-integrin in modulating cardiac aging. Aging Cell 13, 431–440 (2014).
Choi, H. M. T. et al. Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, 1–10 (2018).
bin Imtiaz, M. K. et al. Declining lamin B1 expression mediates age-dependent decreases of hippocampal stem cell activity. Cell Stem Cell 28, 967–977 (2021).
Li, H. et al. Fly Cell Atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. Science https://doi.org/10.1126/science.abk2432 (2022).
Prakash, A. et al. Cardiac abnormalities in patients with Hutchinson–Gilford progeria syndrome. JAMA Cardiol. 3, 326–334 (2018).
Vogler, G. & Ocorr, K. Visualizing the beating heart in Drosophila. J. Vis. Exp. https://doi.org/10.3791/1425 (2009).
Salick, M. R. et al. The scanning gradient Fourier transform (SGFT) method for assessing sarcomere organization and alignment. J. Appl. Phys. 127, 194701 (2020).
Cheedipudi Sirisha, M. et al. Genomic reorganization of lamin-associated domains in cardiac myocytes is associated with differential gene expression and DNA methylation in human dilated cardiomyopathy. Circ. Res. 124, 1198–1213 (2019).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Davidson, E. H. & Erwin, D. H. Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800 (2006).
Schulze, S. R. et al. Molecular genetic analysis of the nested Drosophila melanogaster lamin C gene. Genetics 171, 185–196 (2005).
Brand, A. H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
McGee, M. D. et al. Loss of intestinal nuclei and intestinal integrity in aging C. elegans. Aging Cell 10, 699–710 (2011).
Zhao, M. et al. Segmentation and classification of two-channel C. elegans nucleus-labeled fluorescence images. BMC Bioinformatics https://doi.org/10.1186/s12859-017-1817-3 (2017).
Dreesen, O. et al. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J. Cell Biol. 200, 605–617 (2013).
Schulze, S. R. et al. A comparative study of Drosophila and human A-type lamins. PLoS ONE https://doi.org/10.1371/journal.pone.0007564 (2009).
Cosgrove, B. D. et al. Nuclear envelope wrinkling predicts mesenchymal progenitor cell mechano-response in 2D and 3D microenvironments. Biomaterials 270, 120662 (2021).
Nava, M. M. et al. Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell 181, 800–817 (2020).
Shah, P. et al. Pathogenic LMNA variants disrupt cardiac lamina-chromatin interactions and de-repress alternative fate genes. Cell Stem Cell 28, 938–954.e9 (2021).
Zheng, X. et al. Lamins organize the global three-dimensional genome from the nuclear periphery. Mol. Cell 71, 802–815 (2018).
Hu, B. et al. Plant lamin-like proteins mediate chromatin tethering at the nuclear periphery. Genome Biol. 20, 1–18 (2019).
Ulianov, S. V. et al. Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 10, 1176 (2019).
Sawh, A. N. et al. Lamina-dependent stretching and unconventional chromosome compartments in early C. elegans embryos. Mol. Cell 78, 96–111 (2020).
Chang, L. et al. Nuclear peripheral chromatin-lamin B1 interaction is required for global integrity of chromatin architecture and dynamics in human cells. Protein Cell https://doi.org/10.1007/s13238-020-00794-8 (2020).
Noguchi, A. et al. Decreased lamin B1 levels affect gene positioning and expression in postmitotic neurons. Neurosci. Res. https://doi.org/10.1016/j.neures.2021.05.011 (2021).
Li, C.-L. et al. Region-specific H3K9me3 gain in aged somatic tissues in Caenorhabditis elegans. PLoS Genet. 17, e1009432 (2021).
Pickersgill, H. et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38, 1005–1014 (2006).
van Bemmel, J. G. et al. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS ONE https://doi.org/10.1371/journal.pone.0015013 (2010).
Han, Z. & Olson, E. N. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development 132, 3525–3536 (2005).
Bodyak, N. Gene expression profiling of the aging mouse cardiac myocytes. Nucleic Acids Res. 30, 3788–3794 (2002).
Shen, T. et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J. Clin. Invest. 121, 4640–4654 (2011).
Sakabe, N. J. et al. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum. Mol. Genet. 21, 2194–2204 (2012).
Stennard, F. A. et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development 132, 2451–2462 (2005).
Akazawa, H. & Komuro, I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ. Res. 92, 1079–1088 (2003).
Thattaliyath, B. D., Livi, C. B., Steinhelper, M. E., Toney, G. M. & Firulli, A. B. HAND1 and HAND2 are expressed in the adult-rodent heart and are modulated during cardiac hypertrophy. Biochem. Biophys. Res. Commun. 297, 870–875 (2002).
Natarajan, A. et al. Human eHAND, but not dHAND, is down-regulated in cardiomyopathies. J. Mol. Cell. Cardiol. 33, 1607–1614 (2001).
Bigot, A. et al. Replicative aging down-regulates the myogenic regulatory factors in human myoblasts. Biol. Cell 100, 189–199 (2008).
Musarò, A. et al. Enhanced expression of myogenic regulatory genes in aging skeletal muscle. Exp. Cell. Res. 221, 241–248 (1995).
Smith, M. B., Chaigne, A. & Paluch, E. K. An active contour ImageJ plugin to monitor daughter cell size in 3D during cytokinesis. Methods Cell. Biol. 137, 323–340 (2017).
Beri, P. et al. Cell adhesiveness serves as a biophysical marker for metastatic potential. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-19-1794 (2019).
Ocorr, K., Fink, M., Cammarato, A., Bernstein, S. & Bodmer, R. Semi-automated optical heartbeat analysis of small hearts. J. Vis. Exp. https://doi.org/10.3791/1435 (2009).
Hertz, H. Ueber den kontakt elastischer koerper. J. fuer die Reine Angew. Math. 92, 156 (1881).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Babraham Bioinformatics. FastQC a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
S, A., PT, P. & W, H. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
L, W., S, W. & W, L. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012).
Hennig, C. fpc: Flexible Procedures for Clustering https://cran.r-project.org/web/packages/fpc/index.html (2020).
Alexa, A. & Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.50.0. https://bioconductor.org/packages/release/bioc/html/topGO.html (2022).
Mitchell, A. L. et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360 (2019).
LY, G. et al. The NCBI BioSystems database. Nucleic Acids Res. https://doi.org/10.1093/nar/gkp858 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739 (2011).
A, F. et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018).
DN, S. et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 46, D661–D667 (2018).
Whitehead, A. J., Hocker, J. D., Ren, B. & Engler, A. J. Improved epicardial cardiac fibroblast generation from iPSCs. J. Mol. Cell. Cardiol. 164, 58–68 (2022).
Ahn, K. et al. Selection of internal reference genes for SYBR green qRT–PCR studies of rhesus monkey (Macaca mulatta) tissues. BMC Mol. Biol. 9, 1–8 (2008).
Ruiz-Villalba, A. et al. Reference genes for gene expression studies in the mouse heart. Sci. Rep. 7, 1–9 (2017).
Acknowledgements
The authors thank K. Jepsen of the Institute for Genomic Medicine (UCSD) for technical assistance with sequencing; E. Molina (Sanford Consortium Stem Cell Core) for access and maintenance of the LSM780 microscope; K. Christman, R. Middleton, J. Tuler and K. Birmingham (UCSD) for use and dissection of hearts from the aged mouse model; N. Varki and S. Choudhury of the Mouse Phenotyping Core (UCSD) for assistance with histological preparations; and UCSD undergraduate students S. Skalak, M. Naguib, I. Madan, J. Yun and S. Sharma for assistance with fly husbandry and immunostaining experiments. We thank the Bloomington Drosophila Stock Center (P40OD018537), R. Bodmer, K. Ocorr, J. Grosshans and L. Walrath for providing fly strains. The authors acknowledge funding and equipment support from the NIH (R01AG045428 to A.J.E. and S10OD026929 to UCSD IGM Genomics Center) as well as intramural support from the NIA (to E.G.L. and M.W.). Fellowship support was provided by the American Heart Association (20POST35180048 to N.J.K.), the National Science Foundation (to A.J.W. and P.B.), the ARCS Foundation (to A.J.W.) and the NIH (T32GM008666 to J.D.H.). Work at the Center for Epigenomics was supported in part by the UCSD School of Medicine.
Author information
Authors and Affiliations
Contributions
N.J.K., R.B. and A.J.E. conceived the project and designed experiments. N.J.K. and S.H.S. performed experiments. B.H. and G.V. developed the HCR protocol for adult fly hearts. N.J.K. analyzed experimental data. P.B. developed the Python Lamin distribution analysis pipeline. N.J.K., A.J.W. and J.D.H. performed ATAC-seq and analyzed sequencing data. B.R. oversaw ATAC-seq assay design and provided technical expertise. G.V. oversaw and provided technical expertise for fly imaging and HCR experiments. M.W. performed macaque husbandry and isolated macaque tissue. E.G.L. managed the macaque colony. The manuscript was written by N.J.K., R.B. and A.J.E. with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Nuclear Dynamics in Cardiac and Skeletal Muscle Cells.
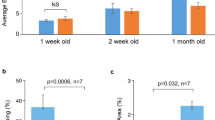
(A) Kaplan–Meier survival curve for yw (gray) and w1118 (black) flies. n = 103 and flies, respectively, were used in the plot. p < 10−3 based on Log-Rank (Mantel-Cox) test between the two strains. (B) Plots of 2D projected area (left) and circularity data (right) for yw flies. n = 129, 108, and 143 for yw flies at 1-, 3-, and 5 weeks, respectively. (C) Cardiomyocyte nuclear area (left) and aspect ratio (right) plotted for w1118 and yw flies as a function of adult age. n = 96, 116, and 141 nuclei for for w1118 flies and n = 129, 108, and 143 for yw flies at 1-, 3-, and 5 weeks of adulthood, respectively. (D) Ventral muscle nuclear area (left), perimeter (center), and aspect ratio (right) from w1118 flies at 1-, 3-, and 5 weeks of adulthood. n = 528, 604, 661 ventral muscle nuclei from w1118 flies at 1-, 3-, and 5 weeks of adulthood. (E) Representative images of the 3D wireframe mesh of cardiomyocyte nuclei from w1118 flies at 1- (top), 3- (middle), and 5 weeks (bottom) of adulthood. Scale bar is 5 μm. *p < 0.05, **p < 10−2, ***p < 10−3, and ****p < 10-4 by one-way ANOVA with Tukey multiple comparisons test. Error bars in all graphs refer to mean + /- SD.
Extended Data Fig. 2 Natural Aging Downregulates LamC and LamB but does not affect their localization.
(A) MA plot of all genes from 1- and 5-week adult w1118 fly hearts showing log2 fold change (FC) and mean normalized expression counts. Data is shown in black for genes −1.25 < FC < 1.25 (dashed lines) or p-adj > 0.05. Open circles represent genes that do not map to a nuclear ontological term. Green and purple data represent DEGs that are up- or downregulated in 5-week adult flies, respectively. (B) Cellular component and molecular function ontological terms for genes associated with the nuclear envelope GO term, organized by their elimination pruning p-value. (C) Representative images of 1 and 5 week w1118 fly heart nuclei stained for DNA, and LamC (green) and LamB (purple) mRNA transcripts. Scale bar, 5 μm. (D) Left plot shows area percentage occupied by mRNA transcripts per cardiomyocyte in 1- and 5-week-old adult w1118 fly hearts. Right plot normalizes data to mean area at 1 week for each transcript. n = 49, 36, 49, and 36 nuclei from w1118 flies at 1 and 5 weeks for LamC and LamB, respectively. (E) Corrected total nuclear fluorescence (CTNF) of 1, 3, and 5 week yw flies for LamC (top) and LamB (bottom). n = 94, 106, and 133 nuclei for LamC and n = 173, 115, and 90 nuclei for LamB for 1, 3, and 5 week adults, respectively. (F) Image showing a representative nucleus with multiple lines radiating out from its centroid (left) to create line plots that are averaged into a single radial profile of the fluorescent intensity (right). Lower panel, the ratio of edge to center intensity is plotted. n = 72, 63, 73, and 33 nuclei from w1118 flies at 1 and 5 weeks for LamC and LamB, respectively. n = 51, 87, 77, and 42 nuclei from yw flies at 1 and 5 weeks for LamC and LamB, respectively. *p < 0.05, **p < 10−2, ***p < 10−3, and ****p < 10-4 by one-way ANOVA with Tukey multiple comparisons test. Bars in (D) refer to min to max, with median and 25th and 75th interquartile range and error bars in (E-F) refer to mean + /- SD.
Extended Data Fig. 3 Validation and Morphological and Functional Characterization of LamB and LamC RNAi lines.
Corrected total nuclear fluorescence (CTNF) for cardiomyocytes from (A) LamC RNAi and (B) LamB RNAi fly lines and their respective controls, that is, attp2 and attp40, n = 47, 36, 79, and 69 nuclei from attp2 and LamC RNAi flies at 1 and 4 weeks (left to right). n = 53, 29, 65, and 61 nuclei from attp40 and LamB RNAi flies at 1 and 4 weeks (left to right). (C) Plots quantifying nuclear perimeter (left, n = 111, 96, 46, and 89 nuclei/condition) and aspect ratio (right, n = 113, 97, 46, and 92 nuclei/condition) for LamB, LamC RNAi and their genetic controls. (D) Plots quantifying nuclear area, perimeter, aspect ratio, and circularity (left to right) for LamB and LamC RNAi lines and their genetic control background at 4 weeks. For all plots, n = 136, 101, 86, and 95 nuclei/condition left to right. (E-F) Plots of diastolic and systolic diameters, and shortening velocity determined from SOHA imaging for attp2 control and LamC RNAi flies (E), and attp40 control and LamB RNAi flies (F) at 1 and 4 weeks. Each LamC RNAi plots, n = 21, 23, 25, and 27 nuclei/condition, left to right. Each LamB RNAi plots, n = 31, 21, 26, and 25 nuclei/condition, left to right. For panels A-D, *p < 0.05, **p < 10−2, ***p < 10−3, and ****p < 10-4 from a two-sided unpaired t-test at each time point and RNAi line. For E-F, *p < 0.05, **p < 10−2, ***p < 10−3, and ****p < 10-4 by one-way ANOVA with Tukey multiple comparisons test. Error bars in (A-D) refer to mean + /- SD, and bars in (E, F) refer to min to max, with median and 25th and 75th interquartile range.
Extended Data Fig. 4 Validation of transgenic fly background, effects of LamB on myogenic transcription factor expression, and LamC heterozygous flies.
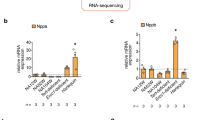
(A) Volcano plot and (B) heat map of bulk RNA-seq from surgically dissected attp2 heart tubes. Fold change represents 5-week attp2 fly hearts normalized to 1-week-old hearts and p-adj was computed from quintuplicate. 1,998 differentially expressed genes (DEGs) were assessed from cutoffs of −0.32 > log2(FC) > 0.32 (or −1.25 > FC > 1.25) (dashed lines) from comparisons of 15 fly hearts per replicate; DEGs increasing and decreasing with age are shown in green and purple, respectively. Heat map columns were hierarchically clustered using Euclidean distance and linkage shown by the dendrogram. Heat maps are normalized within each gene/row. (C) 635 of 688 genes were co-regulated DEGs (92.3%) in the w1118 and attp2 control fly hearts, and plotted based on their fold change with age. DEGs were annotated based on their ontological categorization as nuclear (orange), extracellular matrix (ECM; green), or cytoskeletal (blue). A subset of DEGs either did not fit those categories (black/white) or lacked a known ontology (gray). Only 7.7% of all DEGs were dysregulated. (D) Representative images of attp40 control and LamB RNAi flies at 1 and 4 weeks showing H15, Hand and Tin transcription factor mRNA and DAPI. Scale bar is 5 μm. (E) Quantification of the per cell percentage area for each transcript in attp40 control and LamB RNAi flies at 1 and 4 weeks. For H15, n = 93, 87, 85, and 71 cells, left to right. For Hand, n = 111, 96, 105, and 76 cells, left to right. For Tin, n = 105, 118, 102, and 76 cells, left to right. (F) Quantification of cardiomyocyte nuclear area, circularity and LamC corrected total nuclear fluorescence for 1-week-old female control yw/w1118 and heterozygous LamC excision mutants yw/w1118;LamCex187/+ and yw/w1118;LamC296/+ fly hearts and their representative images in (G) showing staining for LamC (green) and DNA (Magenta), scale bare = 5 µm. For (F), n = 29, 26, 25 (CM nuclei, left to right) (H) Quantification of heart parameters: fractional shortening, diastolic diameter and systolic diameter for background control and heterozygous LamC excision mutants. n = 17, 16, 16 (heart tubes, left to right) (I) Representative images of Hand, H15 and Tin mRNA in heterozygous LamC excision mutants, presented with quantification of per cell percentage area for each transcript in (J). n = 52, 39, 46 (CM nuclei, left to right). *p < 0.05, **p < 10−2, ***p < 10−3, and ****p < 10-4 by one-way ANOVA with Tukey multiple comparisons test. Box plots in (E, H, J) refer to min to max, with median and 25th and 75th interquartile range and error bars in (F) refer to mean + /- SD.
Extended Data Fig. 5 Effects of Adult Myogenic Transcription Factor Loss on of Heart Tube Morphology and Function.
(A) Representative images of A2-A3 heart region visualized by phalloidin (F-Actin) with the indicated transgenes expressed under the control of the Hand-Gal4 promoter. Scale bar is 10 μm. At least three heart tubes were imaged for each condition. (B) Diastolic and (C) systolic diameters of heart tubes from control fly lines (attp40 and attp2) and their corresponding transgenic flies expressing the indicated RNAi. n = 10, 12, 18, 15, 19 and 14 (heart tubes, left to right). (D) CM nuclear area in attp2 control and LamC RNAi hearts subject to regime described in Fig. 6a. n = 29 and 22 (CM Nuclei, left to right). (E-G) Quantification of the per cell, percentage area for Hand, H15 and Tin transcripts upon KD of (E) LamC, (F) Hand (red),Tin (pink) and respective control attp2 (grey), and (G) H15 (blue) and attp40 control (grey), induced as in Fig. 6a. n = 32 and 26 (CM Nuclei/genotype) in (E), n = 20, 23, 28 (CM Nuclei/genotype, left to right) in (F) and n = 43 and 42 (CM Nuclei/genotype) in (G). (H) Schematic of temperature-sensitive transgenic expression where 29 °C enables transgenic expression due to the denaturation of Gal4 transcription factor suppressor, Gal80ts. (I) Fractional shortening of surgically exposed heart tubes at 18 °C and 29 °C for controls (black), and KD of tin (pink), Hand (brown), and H15 (blue) with corresponding diastolic and systolic diameters in (J-M). n = 13, 15, 18, 15, 26, 27, 16, 19, 29, 27, 29, and 21 (heart tubes/transgene/ temperature; left to right as shown in (I)). *p < 0.05, **p < 10−2, ***p < 10−3, and ****p < 10-4 by independent t-test and one-way ANOVA with Kruskal–Wallis test and Dunn’s comparisons test. Box plots show to min to max, with median and 25th and 75th interquartile range and error bars in (D) refer to mean + /- SD.
Extended Data Fig. 6 Effects of LamC and Myogenic Transcription Factor Overexpression.
For panels A-D and as outlined in Fig. 6a, flies were reared at 18 °C and shifted to 25 °C upon eclosure. Nuclear area (A) and CTNF (B) for hearts overexpressing LacZ (control) and LamC from 1 to 7 weeks. n = 31, 41, 23, 30, 26, and 31 cells (A, left to right) and n = 31, 31, 24, 29, 30, and 40 cells (B, left to right). (C) Diastolic and (D) systolic diameters of LacZ and Hand OE flies at 1, 4, and 7 weeks of adulthood. (E) Representative images (left) for nuclei from flies at 18 °C and 29 °C expressing GFPNLS transgene or LamC OE from HandGal,tubGal80ts;tubGal80ts based upon regime described in S5H. Scale bar is 5 μm. Plots of (F) projected nuclear area, (G) circularity, and (H) and corrected total nuclear fluorescence (CTNF) of LamC protein as a function of temperature and transgene expression. n = 39, 40, 46, and 41 nuclei (left to right). (I) For GFPNLS and LamC OE at 18 °C and 29 °C, representative images of LamC mRNA transcripts. Scale bar is 5 μm. (J) Quantification of LamC transcript area per CM in GFPNLS and LamC OE hearts, at 18 °C and 29 °C. n = 24, 36, 20 and 40 CMs, left to right. Diastolic (K) and systolic (L) diameters, and (M) fractional shortening for GFPNLS and LamC OE at 18 °C and 29 °C. n = 32, 19, 30 and 26 heart tubes, left to right. (N). A plot for each transcription factor is shown and quantifies the per cell, percentage area for each transcript. For all transcripts, n = 50, 56, 49 and 35 cells, left to right. (O) Representative images for tin (pink), Hand (brown), and H15 (blue) transcripts in GFPNLS and LamC OE at 18 °C and 29 °C. Scale bar is 5 μm. *p < 0.05, **p < 10−2, ***p < 10−3, and ****p < 10-4 by two-way ANOVA with Sidaks multiple comparisons test (A-D) and one-way ANOVA with Kruskal-Wallis test and Dunn’s comparisons test (F-N). Error bars in (A, B, F-H) refer to mean + /- SD and all box plots display min to max, with median and 25th and 75th interquartile range.
Extended Data Fig. 7 Myogenic transcription factor expression in the left ventricles of aged nonhuman primates and mice.
(A) Expression of three housekeeping genes for mice are plotted as a function of age with a linear and associated p-value shown. Data is plotted for raw Cq values. (B) Expression of four transcription factors in mice is shown, normalized to each housekeeper gene. Data normalized to Eef1e1 (black), Rpl4 (medium gray), and ACTB (light gray) are plotted using the left and right y-axes, depending on the axis label colors. P-values for each fit are shown in the upper right corner. (C) Expression of three housekeeping genes for rhesus macaques are plotted as a function of age with a linear and associated p-value shown. Data is plotted for raw Cq values. (D) Expression of four transcription factors in rhesus macaques is shown, normalized to each housekeeper gene. Data normalized to Rpl13a (black) and TUBB2 (light gray) use the left y-axis whereas Rpl32 uses the right y-axis (medium gray). P-values for each fit are shown in the upper right corner.
Supplementary information
Supplementary Table 1
Age-associated transcriptome changes of w1118 flies. Table showing changes in the transcriptome of w1118 flies. Data listed are annotated for common gene name followed by (left to right): gene description, fold change for the comparison of 5-week/1 week adults, log2FC value, P value, Padj, average log2 expression, raw fragments per kilobase of transcript per million mapped reads (FPKM), gene ID and any gene aliases, if available.
Supplementary Table 2
Gene Ontologies of age-associated transcriptome changes of w1118 flies. Table showing GO associated with transcriptome changes in w1118 flies. Data listed are annotated for (left to right): GO identifier, common name, P value, FDR-adjusted P value, Elim pruning P value, number of genes in term, number of genes that are also in this filter or cluster, number of upregulated genes, number of downregulated genes and the negative log10 Elim pruning P value.
Supplementary Table 3
Differentially accessible chromatin regions for age, LamC RNAi and LamB RNAi. This table annotates ATAC-sequencing data for age comparisons in the w1118 background, in the LamB RNAi versus attp40 flies, and in the LamC RNAi versus attp2 flies. In each tab, data annotates (left to right): peak number, chromosome, peak start location, peak end location, peak annotation, detailed peak annotation, distance to transcription start site (TSS), nearest promoter ID, Entrez ID of nearest gene, Unigene ID for nearest gene, nearest Refseq, gene name, gene alias, gene description, gene type, gene concentration, conc_group1, conc_group2, fold change, P value and FDR.
Supplementary Table 4
Analysis of DARs common to aging versus LamC RNAi and aging versus LamB RNAi. This table annotates co-regulated DAR from ATAC-sequencing data for LamB RNAi versus attp40 flies and for LamC RNAi versus attp2 flies. In tabs one (Age_and_LamC_EdgeR) and three (Age_and_LamB_EdgeR), each comparison is denoted with ‘x’ and ‘y’ corresponding to RNAi versus control and 5wk versus 1wk aging flies, respectively, and annotated for (left to right): peak number, chromosome, peak start location, peak end location, peak annotation, detailed peak annotation, distance to TSS, nearest promoter ID, Entrez ID of nearest gene, Unigene ID for nearest gene, nearest Refseq, gene name, gene alias, gene description, gene type, gene concentration, conc_group1, conc_group2, fold change, P value and FDR. In tabs two (age and LamC genes only) and four (age and LamB genes only), data are annotated just for the fold change for each gene associated with the DAR.
Supplementary Table 5
Gene Ontologies for co-downregulated DAR in aging and LamC iR. This table annotates the GO for biological process (BP) and cellular component (CC) for co-downregulated genes based on the closest assigned gene for the peak and associated DAR. Data are annotated (left to right) for: GO complete name, D. melanogaster peak, number of genes in dataset, expected gene number, gene over/under, fold enrichment, raw P value, FDR and −log10 (FDR P value).
Supplementary Table 6
LamC RNAi-mediated transcriptome changes in attp2 fly background. Table showing changes in the transcriptome of LamC RNAi flies at 1 week of adulthood versus their control attp2 background flies. Data plotted are annotated for common gene name followed by (left to right): gene description, fold change for the comparison of LamC/attp2, log2FC value, P value, Padj, average log2 expression, raw FPKM, gene ID and any gene aliases, if available.
Supplementary Table 7
Age-associated transcriptome changes of attp2 flies. Table showing changes in the transcriptome of attp2 flies. Data plotted are annotated for common gene name followed by (left to right): gene description, fold change for the comparison of 5-week-old/1-week-old adults, log2FC, P value, Padj, average log2 expression, raw FPKM, gene ID and any gene aliases, if available.
Supplementary Table 8
Co-regulated transcriptome changes in attp2 versuss w1118 fly background. Table showing DEGs as a function of age between the attp2 and w1118 fly genotypes. Only genes differentially expressed with age in both systems are shown. Data are annotated for common gene symbol followed by (left to right): gene ID, fly base alias, gene description and then in the attp2 fly first and w1118 second, we show the log2FC value with age, P value for the age fold change and Padj value for the age fold change.
Supplementary Table 9
Co-regulated transcriptome changes in LamC RNAi versus attp2 and biological process ontology terms. Mutually significant DEGs from LamC RNAi and aged fly hearts are listed in this table. In the first tab listing co-regulated genes, data are annotated (left to right) by: symbol, gene ID, gene alias, gene description, base mean gene expression with age, log2FC with age, Padj for age fold change, base mean gene expression for LamC RNAi versus attp2, log2FC for LamC RNAi versus attp2, Padj for LamC RNAi versus attp2 fold change, protein type, additional protein description, protein classification and the type of cellular component GO terms associated with the gene, for example, cytoskeleton, nucleus, extracellular matrix, other or unknown. In the second tab, PANTHER biological process GO terms are annotated for co-regulated terms.
Supplementary Table 10
Primer sequences. All forward and reverse primer sequences for mice and rhesus macaques are shown here.
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 5
Statistical Source Data.
Source Data Fig. 6
Statistical Source Data.
Source Data Fig. 7
Statistical Source Data.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 3
Statistical Source Data.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 6
Statistical Source Data.
Source Data Extended Data Fig. 7
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kirkland, N.J., Skalak, S.H., Whitehead, A.J. et al. Age-dependent Lamin changes induce cardiac dysfunction via dysregulation of cardiac transcriptional programs. Nat Aging 3, 17–33 (2023). https://doi.org/10.1038/s43587-022-00323-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-022-00323-8
This article is cited by
-
Quercetin and dasatinib, two powerful senolytics in age-related cardiovascular disease
Biogerontology (2024)
-
Nuclear remodelling contributes to cardiac dysfunction during ageing
Nature Reviews Cardiology (2023)