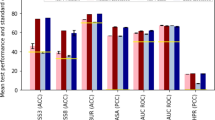
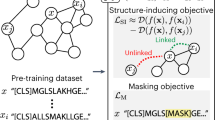
Abstract
Pretrained language models have shown promise in analysing nucleotide sequences, yet a versatile model excelling across diverse tasks with a single pretrained weight set remains elusive. Here we introduce RNAErnie, an RNA-focused pretrained model built upon the transformer architecture, employing two simple yet effective strategies. First, RNAErnie enhances pretraining by incorporating RNA motifs as biological priors and introducing motif-level random masking in addition to masked language modelling at base/subsequence levels. It also tokenizes RNA types (for example, miRNA, lnRNA) as stop words, appending them to sequences during pretraining. Second, subject to out-of-distribution tasks with RNA sequences not seen during the pretraining phase, RNAErnie proposes a type-guided fine-tuning strategy that first predicts possible RNA types using an RNA sequence and then appends the predicted type to the tail of sequence to refine feature embedding in a post hoc way. Our extensive evaluation across seven datasets and five tasks demonstrates the superiority of RNAErnie in both supervised and unsupervised learning. It surpasses baselines with up to 1.8% higher accuracy in classification, 2.2% greater accuracy in interaction prediction and 3.3% improved F1 score in structure prediction, showcasing its robustness and adaptability with a unified pretrained foundation.
Similar content being viewed by others
Main
RNA is a critical molecule in the central dogma of molecular biology, which describes the flow of genetic information from DNA to RNA to protein.
RNA molecules play a crucial role in various cellular processes, including gene expression, regulation and catalysis. Given the importance of RNA in biological systems, there is a growing demand for efficient and accurate methods to analyse RNA sequences. The analysis of RNA sequences has traditionally been performed using experimental techniques such as RNA sequencing and microarrays1,2. However, these methods are often expensive and time-consuming and require large amounts of input RNA. In recent years, there has been increasing interest in using computational methods based on machine learning models to analyse RNA sequences.
Pretrained language models, on the other hand, have shown great success in various natural language processing tasks, including text classification3, question answering4 and language translation5. Advancements in the field of natural language processing have led to the successful adoption of pretrained language models like BERT6 to model and analyse nucleotides (nts) and ribonucleotides from trillions of DNA/RNA sequences. For example, preMLI7 employs rna2vec to produce RNA word vector representations. The RNA sequence features are then mined independently, and the two feature vectors are concatenated as the input for the prediction task. DNABERT8 has been proposed to extract features from DNA sequences via the pretrained language model BERT-alike, and its derivatives9,10 with task-agnostic extensions have been studied to solve DNA analytical tasks in an ad hoc manner11. Moreover, based on T5 (ref. 12), Rm-LR13 integrates two large-scale RNA language pretrained models to learn local key features and collect discriminative sequential information. A bilinear attention network is then used to integrate the learned features. However, there is still some work focusing on generic models that performs well on varying downstream tasks derived from one set of pretrained weights. RNA-FM14 trains a foundation model for the community to fit all the ncRNA sequences, although it only uses naive token masking as a pretraining strategy, which may lose high-density information hidden in continuous RNA subsequences. This problem is further compounded by the fact that RNA is a more complex molecule than DNA15, due to the presence of additional modifications and higher-order structures, and existing pretrained models are not optimized for RNA analysis.
In response to this challenge, we have developed a pretrained RNA language model: RNAErnie. As shown in Fig. 1, this model is built upon the Enhanced Representation through Knowledge Integration (ERNIE) framework and incorporates multilayer and multihead transformer blocks, each having a hidden state dimension of 768. Pretraining is conducted using an extensive corpus consisting of approximately 23 million RNA sequences meticulously curated from RNAcentral16. The proposed motif-aware pretraining strategy involves base-level masking, subsequence-level masking and motif-level random masking, which effectively captures both subsequence and motif-level knowledge17,18,19, enriching the representation of RNA sequences as illustrated in Fig. 2a. Additionally, RNAErnie tokenizes coarse-grained RNA types as special vocabularies and appends the tokens of coarse-grained RNA types at the end of every RNA sequence during pretraining. By doing so, the model gains the potential to discern the distinct characteristics of various RNA types, facilitating domain adaption to various downstream tasks.
The RNAErnie model consists of 12 transformer layers. In the motif-aware pretraining phase, RNAErnie is trained on a dataset of approximately 23 million sequences extracted from the RNAcentral database using self-supervised learning with motif-aware multilevel random masking. In the type-guided fine-tuning phase, RNAErnie first predicts the possible coarse-grained RNA types using output embeddings and then leverages the predicted types as auxiliary information for fine-tuning the model with task-specific heads. w/, with; w/o, without; MLM, masked language modelling; norm., normalization.
a, Motif-aware multilevel masking strategy for RNAErnie pretraining. Built upon ERNIE transformer blocks, the design incorporates three levels of masking: base, subsequence and motif. All masking levels are applied during the pretraining phase. b, Type-guided fine-tuning for downstream tasks. The RNAErnie approach first leverages an RNAErnie basic block to predict the top-K most possible coarse-grained RNA types. Then, it stacks an additional layer of K downstream modules with shared parameters for fine-tuning and outputs the ensemble result for tasks.
Specifically, a type-guided fine-tuning strategy is employed, incorporating the predicted RNA types as ‘auxiliary information’ within a stacking architecture, as shown in Fig. 2b. Upon receiving an RNA sequence as input, the model first employs a pretrained RNAErnie block to generate output embeddings. Subsequently, it predicts the potential coarse-grained RNA types based on these embeddings. The sequence and the predicted RNA types are then fed into a downstream network, which consists of RNAErnie blocks and task-specific heads. This approach enables the model to accommodate a diverse range of RNA types and enhances its utility in a broad spectrum of RNA analytical tasks. More specifically, to adapt the distribution shifts between pretraining datasets and target domains, RNAErnie leverages domain adaptation20 that composites the pretrained backbone with downstream modules in three neural architectures: frozen backbone with trainable head (FBTH), trainable backbone with trainable head (TBTH) and stacking for type-guided fine-tuning (STACK). In this way, the proposed method can either end-to-end optimize the backbone and task-specific heads or fine-tune task-specific heads with embeddings extracted from the frozen backbone, subject to the downstream applications.
The conducted experiments highlight the immense potential of RNAErnie in advancing RNA analysis. The model demonstrates strong performance across diverse downstream tasks, showcasing its versatility and effectiveness as a generic solution. Additionally, the innovative strategies employed in RNAErnie show promise in enhancing the performance of other pretrained models in RNA analysis. These findings position RNAErnie as a valuable asset, empowering researchers with a powerful tool to unravel the complexities of RNA-related investigations.
Results
In this section, we present the experiment results for RNAErnie evaluation on both unsupervised learning (RNA grouping) and supervised learning (RNA sequence classification, RNA–RNA interaction prediction and RNA secondary structure prediction) tasks. For additional experiment settings and results (such as long-sequence classification, SARS-CoV-2 variant evolutionary path visualization and so on), please refer to Supplementary Information Section C.
Unsupervised clustering of RNAErnie-extracted features
Various types of RNA exhibit distinct functions and structures, and it is expected that these characteristics are captured within the embeddings generated by our proposed model (RNAErnie) using raw RNA sequences. To examine the patterns within the known RNA repertoire, we utilize the suggested encoder to establish scatter plots of RNA sequences. Dimension reduction using PHATE21 is then employed to map the embeddings onto a two-dimensional plane. We evaluate the impact of the learning process by considering both pretrained and randomly initialized RNAErnie embeddings, as well as 3mer statistical embeddings22 for visualization.
Figure 3a shows the results, where the pretrained RNAErnie embedding space effectively organizes RNA types into distinct clusters based on their structural and functional properties. We also use a random model for comparing encoding effects, establishing a baseline for comparison with other encoding methods. This comparison allows us to evaluate the effectiveness of each method in enhancing the encoding process. The random model exhibits a less-defined clustering structure, and the 3mer embeddings lack distinguishable features. This indicates that RNAErnie captures structural and functional information beyond the primary structure of RNA, enabling grouping based on similar properties. To investigate the diversity of non-coding RNAs (ncRNAs), we categorize them using sequence ontology at various levels. Figure 3b illustrates selected classes of ncRNA, such as ribosomal RNA (rRNA), long ncRNA (lncRNA) and small ncRNA (sncRNA). Figure 3c shows the high-level ontology relationships between ncRNA, transcript, messenger RNA (mRNA) and intron RNA. Figure 3d represents the low-level ontology of small regulatory ncRNA. RNAErnie effectively discriminates between classes at different ontology levels, while the 3mer statistical embeddings struggle to separate them. This suggests that RNAErnie captures structural or functional similarities rather than relying solely on the length of ncRNAs. Note that the random approach seems to outperform RNAErnie in differentiating between classes across various ontology levels. This finding suggests that RNAErnie might be less effective in capturing the ontology patterns of low-level, small regulatory ncRNA classes. We believe that this limitation in identifying low-level ontology patterns may stem from several factors, including the complexity and heterogeneity of classes at this level or potential biases in our training dataset. Further research and detailed analysis are needed to identify the specific causes behind RNAErnie’s reduced efficacy in discerning patterns in low-level ontology.
Left, RNAErnie embeddings. Middle, randomly initialized embeddings. Right, 3mer statistical embeddings. a, Scatter plot showcasing all ncRNA types within a subset of RNAcentral, utilizing different embedding methods. b, Embedding projections of selected ncRNA classes, including rRNA, lncRNA, RNase P RNA, ribozyme, sncRNA, signal recognition particle RNA (SRP RNA), antisense RNA and Y RNA. c, Distribution of embeddings based on high-level sequence ontology, encompassing ncRNA, transcript, mRNA and intron RNA. d, Detailed distribution of embeddings for low-level small regulatory ncRNA classes, such as Piwi-interacting RNA (piRNA), miRNA, transfer-messenger RNA (tmRNA) and small interfering RNA (siRNA).
In total, these findings demonstrate that RNAErnie constructs scatter plots by capturing the structural and functional characteristics of ncRNAs, going beyond nucleic acid statistics alone.
Supervised domain adaptation on downstream tasks
In this section, we demonstrate the effectiveness of RNAErnie in three essential supervised learning tasks: RNA sequence classification, RNA–RNA interaction and RNA secondary structure prediction.
To reveal the effectiveness of the designs in RNAErnie, we conducted a series of ablation studies using variant models derived from RNAErnie. These models vary in complexity, beginning with Ernie-base, which lacks RNA-specific pretraining and includes standard fine-tuning. RNAErnie−− employs base-level masking during pretraining, and RNAErnie− adds subsequence-level masking to the mix. The complete RNAErnie model further integrates motif-level masking and is fine-tuned using either TBTH or FBTH architectures. Extending this, RNAErnie+ represents the apogee of complexity within this family, including all three levels of masking and a STACK architecture for pretraining. Lastly, the RNAErnie without chunk model is tailored for long RNA sequences by truncating and discarding segments to contend with computational constraints, aimed at the efficient classification of long non-coding and protein-encoding transcripts.
In addition, we also bring pretrained models from existing literature, including RNABERT23, RNA-MSM24 and RNA-FM14 for comparison.
RNA sequence classification
We evaluate the performance of our proposed sequence-classification models on the benchmark nRC25. This dataset consists of ncRNA sequences selected from the Rfam database release 12 (ref. 26). nRC is composed of a balanced collection of sequences, with 20% non-redundant samples for each of the 13 classes. It has 6,320 training sequences and 2,600 testing sequences labelled with 13 classes.
Table 1 presents the sequence-classification results for RNAErnie on the nRC dataset. The table includes several baseline methods as well as different variants of the RNAErnie models. The baseline values are all taken from cited literature except the pretrained models: RNABERT, RNA-MSM and RNA-FM. Analysing the performance of the models, we observe that the baseline methods achieve varying levels of accuracy. Notably, ncRDense demonstrates decent performance, achieving high accuracy, recall, precision, F1 score and Matthews correlation coefficient (MCC) values. Turning our attention to the RNAErnie variants, we can see that they consistently outperform most of the baseline models across all evaluation metrics. Although ncRDense can beat the first two (that is, Ernie-base and RNAErnie−−), RNAErnie−, RNAErnie and RNAErnie+ show better performance in all five dimensions.
In the hierarchy of the RNAErnie model family, performance metrics improve incrementally with complexity of design. The foundational model, Ernie-base, establishes a baseline that is modestly surpassed by RNAErnie−− through the introduction of base-level masking in pretraining. Furthermore, RNAErnie− incorporates subsequence-level masking and delivers notably enhanced accuracy, recall, precision, F1 score and MCC values, endorsing the value of a more comprehensive masking strategy. The full RNAErnie model integrates base, subsequence and motif-level masking, achieving superior performance over its predecessors across all metrics and illustrating the cumulative benefits of multilevel masking. The apex model, RNAErnie+, which employs an exhaustive masking regimen in conjunction with a two-stage fine-tuning architecture, outperforms all variants in our experiments.
RNA–RNA interaction
We evaluate the performance of our model on one of the most representative benchmark datasets, DeepMirTar27,28, which is used for predicting the interaction between microRNAs (miRNAs) and mRNAs. This dataset consists of 13,860 positive pairs and 13,860 negative pairs. The miRNA sequences in DeepMirTar are all shorter than 26 nts, and the mRNA sequences are shorter than 53 nts. Because most of the target sites are believed to be located at the 3′ untranslated region, DeepMirTar only considers them. Furthermore, two seeds were taken into consideration: the non-canonical seed, which pairs at position 2-7 or 3-8, permitting G-U couplings and up to one bulged or mismatched nt; and the canonical seed, which is the precise W-C pairing of 2-7 or 3-8 nts of the miRNA. Given that RNA types (miRNA, mRNA) are fixed here, we do not test RNAErnie+ version which uses a two-stage pipeline here.
Table 2 presents the performance comparison between the proposed RNAErnie models and baseline methods from existing literature, such as Miranda29, RNAhybrid30, PITA31, TargetScan v.7.0 (ref. 32), TarPmiR33 and DeepMirTar27. The baseline values are all taken from cited literature except the pretrained models: RNABERT, RNA-MSM and RNA-FM. These are evaluated on the RNA–RNA interaction prediction task using the DeepMirTar dataset. DeepMirTar emerges as a strong baseline, exhibiting high scores across all metrics. The Ernie-base model and the RNAErnie variations are then assessed, with the RNAErnie model demonstrating superior performance and particularly excelling in accuracy, precision, F1 score and area under the curve (AUC). This variation achieves an impressive accuracy score of 0.9872, a competitive precision score of 0.9901, an F1 score of 0.9873 and the highest AUC score of 0.9976, indicating excellent overall performance and discriminative power.
Overall, the results suggest that the RNAErnie model, particularly the RNAErnie variation, outperforms the existing methods and the Ernie-base model in the RNA–RNA interaction prediction task. These findings highlight the potential of the RNAErnie model in accurately predicting RNA–RNA interactions.
RNA secondary structure prediction
This section presents a comprehensive comparison between our pretrained RNAErnie model and several baseline models, including the state-of-the-art UFold model34, in the context of RNA secondary structure prediction tasks. The experiments are conducted using commonly used benchmarks employed in state-of-the-art models. These benchmarks include:
-
RNAStralign35: This dataset comprises 37,149 RNA structures from eight RNA families, with lengths ranging from approximately 100 to 3,000 base pairs (bp).
-
ArchiveII36: This dataset consists of 3,975 RNA structures from ten RNA families, with lengths ranging from approximately 100 to 2,000 bp.
-
bpRNA-1m37: This dataset contains 13,419 RNA structures from 2,588 RNA families, with sequence similarity removed using an 80% sequence-identity cut-off. The lengths of the sequences range from approximately 100 to 500 bp. The dataset is randomly split into three subsets: TR0 (10,814 structures) for training, TV0 (1,300 structures) for validation and TS0 (1,305 structures) for testing.
We train our model on the entire RNAStralign dataset, as well as the TR0 subset and other augmented mutated datasets, following the approach used in UFold. Subsequently, we evaluate performance on the ArchiveII600 dataset, which is a subset of ArchiveII with lengths less than 600 bp, and the TS0 dataset.
Table 3 presents a comparative analysis of the performance of various methods on the RNA secondary structure prediction task using the ArchiveII and TS0 datasets. The table presents the results of several baseline methods, including RNAstructure, RNAsoft, RNAfold, MXfold2, Mfold, LinearFold, Eternafold, E2Efold, Contrafold and Contextfold. Each method is assessed based on its precision, recall and F1 score for both the ArchiveII600 and TS0 datasets. The baseline values are all taken from cited literature except the pretrained models: RNABERT, RNA-MSM and RNA-FM. Among the RNAErnie variations, RNAErnie+ achieves the highest scores in precision, recall and F1 score, indicating its superior performance in RNA secondary structure prediction. Notably, RNAErnie+ achieves a remarkable precision score of 0.886, a high recall score of 0.870 and an impressive F1 score of 0.875 on the ArchiveII600 dataset. These results highlight the effectiveness of RNAErnie+ in accurately predicting RNA secondary structures.
Discussion
Our method, RNAErnie, outperforms existing advanced techniques across seven RNA sequence datasets encompassing over 17,000 major RNA motifs, 20 RNA classes/types and 50,000 RNA sequences. Evaluation using 30 mainstream RNA sequence technologies confirms the generalization and robustness of RNAErnie. We employed accuracy, precision, recall, F1 score, MCC and AUC as evaluation metrics to ensure a fair comparison of RNA sequence-analysis methods. Currently, little research exists on applying transformer architectures with enhanced external knowledge to RNA sequence data analysis. Our from-scratch RNAErnie framework integrates RNA sequence embedding and a self-supervised learning strategy, resulting in superior performance, interpretability and generalization potential for downstream RNA tasks. Additionally, RNAErnie is adaptable to other tasks through modification of the output and supervision signals. RNAErnie is publicly available and serves as an effective tool for understanding type-guided RNA analysis and advanced applications.
The RNAErnie model, despite its innovations in RNA sequence analysis, confronts several challenges. First, the model is constrained by the size of the RNA sequences it can analyse, as sequences longer than 512 nts are dropped, potentially omitting vital structural and functional information. The chunking method developed to handle longer sequences might result in the further loss of information about long-range interactions. Second, the focus of this study is narrow, centred only on the RNA domain and not extending to tasks like RNA-protein prediction or binding-site identification. Additionally, the model encounters difficulties in considering three-dimensional structural motifs of RNAs, such as loops and junctions, which are essential for understanding RNA functions.
More importantly, the existing post hoc architectural design has potential limitations, including heightened inference overhead. An alternative approach involves designing a specialized loss function that incorporates RNA type information and pretraining the model in an end-to-end fashion. We have experimented with this concept and engaged in preliminary pretraining. Our findings indicate that although this method proves beneficial for discriminative tasks such as sequence classification, it unfortunately leads to suboptimal token representations with performance degradation in reconstruction of structures. Detailed information is provided Supplementary Information Section C.6. Our future work will go deeper into this issue and explore solutions.
Methods
This section provides a comprehensive overview of the design features associated with each component of RNAErnie. We will explore the specific characteristics of each element and discuss their collaborative functionality in enabling the accomplishment of diverse downstream tasks.
Overall design
In this work, we present RNAErnie, an approach for large-scale pretraining of RNA sequences based on the ERNIE framework38, which incorporates multilayer and multihead transformer blocks39.
RNAErnie transformer
The basic block of the RNAErnie transformer shares the same architectural configuration as ERNIE38, employing a 12-layer transformer and a hidden state dimension of Dh = 768. Consider an input RNA sequence denoted as x = (x1, x2, ⋯ , xL), where each element xi ∈ {‘A’, ‘U’, ‘C’, ‘G’} and L represents the length of the sequence. An RNAErnie block first tokenizes RNA bases in the sequence and subsequently feeds them into the transformer. This process enables us to extract token embeddings \({{{\bf{h}}}}=({h}_{1},{h}_{2},\cdots \,,{h}_{L})\in {{\mathbb{R}}}^{L\times {D}_{{\mathrm{h}}}}\), where Dh represents the dimension of the hidden representations for the tokens. Given the embeddings for every token in the RNA sequence, the RNAErnie basic block transforms the series of token embeddings into a lower-dimensional vector (that is, 768 dimensions) using trainable parameters38 and then outputs the embedding of the RNA sequence. The total number of trainable parameters in RNAErnie is approximately 105 million.
Pretraining datasets
Basically, like many other pretraining based approaches, the RNAErnie approach is structured into two main phases: pretraining and fine-tuning. In the pretraining phase, which is agnostic to any specific task, RNAErnie is meticulously trained on a vast corpus of 23 million ncRNA sequences obtained from the RNAcentral database16. This self-supervised autoregressive training phase allows RNAErnie to capture sequential distributions and patterns within the RNA sequences, thereby acquiring a comprehensive understanding of their structural and functional information. In the subsequent task-specific fine-tuning phase, the pretrained RNAErnie model is either fine-tuned with downstream modules or used to generate sequence embeddings (features) that complement a lightweight prediction layer. Regarding the tokenization of RNA bases, the sequences are tokenized to represent ‘A’, ‘T/U’, ‘C’ and ‘G’, with the initial token of each sequence reserved for the special classification embedding ([CLS]). Additionally, an indication embedding ([IND]) is appended to each RNA sequence, followed by indication classes (for example, ‘miRNA’, ‘mRNA’, ‘lnRNA’) derived from the RNAcentral database, as depicted in Extended Data Fig. 1. The inclusion of the indication embedding encourages the model to cluster similar RNA sequences in a latent space, facilitating retrieval-based learning40.
Motif-aware pretraining strategies
To integrate both subsequence and motif-level knowledge into the representation of RNA sequences, we introduce a motif-ware multilevel masking strategy to pretrain the RNAErnie basic block, as opposed to directly incorporating motif embedding. In addition, the RNAErnie approach follows the standard routine of pretraining with all three levels of masking tasks, learning to predict the masked tokens and also capture contextualized representations of the input RNA sequence. Specifically, the procedure of RNAErnie pretraining with motif-aware multilevel masking strategies is as follows.
Base-level masking
In the initial stage of the learning process, we employ base-level masking as a crucial component. Specifically, we randomly mask 15% of the nucleobases within an RNA sequence. Among the masked positions, 10% are preserved without any alterations, and the remaining 10% are replaced with other nucleobases. The model takes the remaining nucleobases as input and is tasked with predicting the masked positions. This stage primarily focuses on acquiring fundamental token representations; capturing intricate higher-level biological insights proves to be a challenging endeavour.
Subsequence-level masking
Next, we incorporate the masking of random subsequences, which are short and contiguous segments of nucleobases within an RNA sequence. Previous studies, such as refs. 41 and 42, have demonstrated the efficacy of contiguous token masking in enhancing pretrained models for span-selection tasks. Additionally, it is important to consider that the functionality of nucleobases often manifests within the context of sequential arrangements. By predicting these subsequences as a whole, we encourage the model to capture a deeper understanding of the biological information inherent in the relationships between consecutive nucleobases. In our research, we specifically mask subsequences with lengths ranging from 4 to 8 bp.
Motif-level masking
In the final stage of pretraining, we employ motif-level masking as part of our approach. RNA motifs, characterized as recurrent structural elements with a high concentration of information, have been extensively observed in atomic-resolution RNA structures17. These motifs are widely recognized for their crucial involvement in various biological activities, such as the formation of RNA tertiary structures19, interaction with dsRNA-binding proteins (RBPs) and participation in complex formation with proteins18. To incorporate these motifs into our model as so-called biological priors, we gather them from multiple sources:
-
ATtRACT43: This resource provides comprehensive information on 370 RBPs and 1,583 RBP consensus binding motifs. The data is extracted and carefully curated from experimentally validated sources such as CISBP-RNA, SpliceAid-F and RBPDB databases.
-
SpliceAid44: We gather information from SpliceAid, which encompasses 2,220 target sites associated with 62 human splicing proteins. Additionally, it includes expression data from 320 tissues per cell.
-
We also extract the most frequently occurring contiguous nucleobase sequences, ranging from 4 to 8 bp, by scanning the entirety of the RNAcentral database.
By incorporating motifs from these diverse sources, we aim to capture a comprehensive representation of RNA structural elements for our analysis.
Type-guided fine-tuning strategy
Given the RNAErnie basic block pretrained with motif-aware multilevel masking strategies, we need to combine the basic blocks of the RNAErnie transformer with task-specific heads—for example, a fully connected layer for RNA classification—into a neural network for the downstream task and further train the neural network subject to labelled datasets for the downstream application in a supervised learning manner. Here, we introduce our proposed type-guided fine-tuning strategy in two parts: neural architectures for tasks and domain-adaptation strategies.
Neural architectures for fine-tuning
To adapt various downstream tasks, the RNAErnie approach follows the surgical fine-tuning strategies20 and offers three sets of neural network architectures as follows.
FBTH
In the FBTH architecture, given RNA sequences and their labels for a downstream task, the RNAErnie approach simply extracts embeddings of RNA sequences from a pretrained RNAErnie basic block and then leverages the embeddings as inputs to train a separate task-specific head subject to the downstream tasks. In this way, the parameters in the RNAErnie backbone are frozen, while the head is trainable. According to ref. 20, this architecture would work well when the downstream tasks are out-of-distribution of pretraining datasets.
TBTH
In the TBTH architecture, the RNAErnie approach directly combines the RNAErnie basic block and the task-specific head to construct an end-to-end neural network for downstream tasks and then trains the neural network using the labelled datasets in a supervised learning manner. In this way, the parameters in both the RNAErnie backbone and the head are trainable. According to ref. 20, this architecture would work well when the downstream tasks and pretraining datasets are in the same distribution.
STACK
In the STACK architecture, the RNAErnie approach first leverages an RNAErnie basic block to predict the top-K most possible coarse-grained RNA types (that is, the K coarse-grained RNA types with the highest probabilities) using the input RNA sequence. Then it stacks an additional layer of K downstream modules with shared parameters for fine-tuning, where every downstream module refers to a TBTH/FBTH network and is fed with the RNA sequence and a predicted RNA type for the downstream task. The K downstream modules output K prediction results, and the RNAErnie approach outputs the ensemble of K results as the final outcome.
More specifically, in the STACK architecture, the RNAErnie basic block first predicts the indication of an RNA sequence following the [IND] marker by estimating the probability of the masked indication token, denoted as p(xIND∣x; θ). From these predictions, the RNAErnie approach selects the top-K indications, denoted as \({I}_{k}\in {{{\mathcal{I}}}}\) for k = 1, ⋯ , K, along with their corresponding probabilities σ1, …, σK. Each selected indication is then appended to the end of the RNA sequence, resulting in K parallel inputs to the downstream module. Then the downstream module takes the K parallel inputs simultaneously, enabling ensemble learning through soft majority voting. Specifically, the RNAErnie approach calculates the weighted sum for soft majority voting as follows:
where qk could be either scalar, vector or matrix outputs from the downstream module for various downstream tasks (for example, logit vectors for classification tasks or pair-wise feature maps for structural analysis), while \(\bar{q}\) refers to the weight sum.
Note that although we consider the stacking architecture part of our key contributions, FBTH and TBTH sometimes deliver better performance.
Domain adaptation to downstream tasks
Upon completion of the pretraining phase, the RNAErnie basic block is prepared for type-guided fine-tuning, enabling its application to various downstream tasks. It is important to emphasize that RNAErnie has the potential to accommodate a diverse array of tasks, extending beyond the examples provided below, through appropriate FBTH, TBTH and STACK architectures.
RNA sequence classification
RNA sequence classification is a pivotal task that assigns RNA sequences to specific categories. In other words, it maps an RNA sequence x of length L to scalar labels, which refer to different categories. RNA sequence classification is crucial for understanding their functions and their roles in various biological processes. Accurate classification of RNA sequences enables researchers to identify ontology and predict functions, which facilitates the development of new therapies and treatments for RNA-related diseases.
Our work leverages STACK with TBTH to classify RNA sequences. It stacks K classification modules: the RNAErnie basic block combined with a trainable MLP as a prediction head. However, the computational complexity of transformers, which exhibit a quadratic time complexity of \({{{\mathcal{O}}}}({n}^{2}d)\), where n denotes the sequence length, posed challenges when processing excessively long RNA sequences. To discern lncRNA amidst protein-coding transcripts, we employed a chunk strategy. This strategy entails the division of lengthy RNA sequences into more manageable segments, which are independently fed into the RNAErnie approach. Subsequently, we aggregate the segment-level logits to obtain the sequence-level logit and employ an MLP for classification purposes.
RNA–RNA interaction prediction
RNA–RNA interaction prediction refers to the estimation of interactions between two RNA sequences, such as miRNA and mRNA, circular RNA and lncRNA. This task maps two RNA sequences, xa of length L1 and xb of length L2, to binary labels 0/1, where 0 indicates no interaction between the two RNA sequences and 1 indicates interaction. Accurate prediction of RNA–RNA interactions can provide valuable insights into RNA-mediated regulatory mechanisms and enhance our understanding of biological processes, including gene expression, splicing and translation45.
Our work employs a TBTH architecture, which combines the RNAErnie basic block with a hybrid neural network inspired by ref. 46. This hybrid neural network acts as the interaction prediction head, sequentially incorporating several components: a convolutional neural network, a bidirectional long short-term memory network and a MLP. Because the types of interacting RNA are fixed, it is unnecessary to employ the STACK architecture for the purpose of RNA–RNA interaction analysis.
RNA secondary structure prediction
RNA secondary structure prediction determines the probable arrangement of bp within an RNA sequence, which can fold back onto itself and form specific pairings. It maps an RNA sequence x of length L to a 0/1 matrix with shape L × L, where element i, j means whether nt i forms bp with nt j. The secondary structure of RNA plays a critical role in understanding its interactions with other molecules and its functional importance. This prediction technique is a valuable tool in molecular biology, aiding in the identification of potential targets for drug design and enhancing our understanding of gene expression and regulation mechanisms.
Our work utilizes the STACK architecture with FBTH to fold RNA sequences. We combined the RNAErnie basic block with a folding neural network inspired by the methodology described in ref. 47. It computes four distinct folding scores—helix stacking, unpaired region, helix opening and helix closing—for each pair of nt bases. Subsequently, we utilize a Zuker-style dynamic programming approach48 to predict the most favourable secondary structure. This is achieved by maximizing the cumulative scores of adjacent loops, following a systematic and rigorous computational procedure. To facilitate the training of our deep neural network, we adopt the max-margin framework. Within this framework, the network minimizes the structured hinge loss function while incorporating thermodynamic regularization.
Hyperparameters and configurations
During the pretraining phase, our model underwent approximately 2,580,000 steps of training, with a batch size set to 50 and a maximum sequence length for ERNIE limited to 512. We utilized the AdamW optimizer, which was regulated by a learning-rate schedule involving anneal warm-up and decay. The initial learning rate was set at 1 × 10−4, with a minimum learning rate of 5 × 10−5. The learning-rate scheduler was designed to warm up during the first 5% of the steps and then decay in the final 5% of the steps. In terms of masking strategies, we maintained a proportion of 1:1:1 across the three different masking levels, with the training algorithm randomly selecting one strategy for each training session. The pretraining was conducted on four Nvidia Tesla V100 32 GB graphics processing units, taking around 250 hours to reach convergence.
Here, in additional to the hyperparameters for pretraining, we introduce the configurations of variant pretrained models derived from RNAErnie and used in experiments:
-
Ernie-base: this model represents the vanilla ERNIE architecture without any pretraining on RNA sequence datasets. It underwent standard fine-tuning.
-
RNAErnie−−: in this model, only base-level masking was employed during the pretraining phase of the RNAErnie family. It was then fine-tuned using the standard approach.
-
RNAErnie−: the RNAErnie family model with both base and subsequence-level masking during pretraining, followed by standard fine-tuning.
-
RNAErnie: this model encompasses the complete set of masking strategies, including base, subsequence and motif-level masking during pretraining. It was fine-tuned using the TBTH or FBTH architecture.
-
RNAErnie+: the most comprehensive model in the RNAErnie family, incorporating all three levels of masking during pretraining and the STACK architecture.
-
RNAErnie without chunk: this model truncates RNA sequences and discards any remaining segments when classifying long RNA sequences, specifically lncRNA (for example, in lnRC_H and lnRC_M datasets) alongside protein-encoding transcripts.
Data availability
The datasets used for pretraining and fine-tuning are all derived from previous studies. Here we include the official links. Note that the lncRNA_H and lncRNA_M datasets are used for long-sequence classification in the Supplementary Information. RNAcentral16: https://ftp.ebi.ac.uk/pub/databases/RNAcentral/releases/21.0/; ATtRACT43: https://attract.cnic.es/download; SpliceAid44: http://193.206.120.249/cgi-bin/SpliceAid.pl?sites=Download; nRC25: http://tblab.pa.icar.cnr.it/public/nRC/paper_dataset/; lncRNA_H49: https://www.gencodegenes.org/human/release_25.html; lncRNA_M49: https://www.gencodegenes.org/mouse/; DeepMirTar27: https://github.com/tjgu/miTAR/tree/master/scripts_data_models; ArchiveII36: https://rna.urmc.rochester.edu/publications.html; RNAStrAlign35: https://github.com/mxfold/mxfold2/releases/tag/v0.1.0; bpRNA37: https://bprna.cgrb.oregonstate.edu/download.php#bpRNA. Source data are provided with this paper.
Code availability
We built RNAErnie using Python and the PaddlePaddle deep learning framework. The code repository of RNAErnie, readme files and tutorials are all available at ref. 50. A docker image with configured environments and dependent libraries is available for download at ref. 51. To compare pretrained RNA language baselines, see the code repository at ref. 52.
References
Kukurba, K. & Montgomery, S. RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015, pdb–top084970 (2015).
Conesa, A. et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 1–19 (2016).
Dharmadhikari, S., Ingle, M. & Kulkarni, P. Empirical studies on machine learning based text classification algorithms. Adv. Comput. 2, 161 (2011).
Zheng, S., Li, Y., Chen, S., Xu, J. & Yang, Y. Predicting drug-protein interaction using quasi-visual question answering system. Nat. Mach. Intell. 2, 134–140 (2020).
Min, B. et al. Recent advances in natural language processing via large pre-trained language models: a survey. ACM Comput. Surv. 56, 1–40 (2021).
Kenton, J. & Toutanova, L. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (eds Burstein, J. et al.) 4171–4186 (Association for Computational Linguistics, 2019).
Yu, X., Jiang, L., Jin, S., Zeng, X. & Liu, X. preMLI: a pre-trained method to uncover microRNA-lncRNA potential interactions. Brief. Bioinform. 23, bbab470 (2022).
Ji, Y., Zhou, Z., Liu, H. & Davuluri, R. DNABERT: pre-trained bidirectional encoder representations from transformers model for DNA-language in genome. Bioinformatics 37, 2112–2120 (2021).
Leksono, M. & Purwarianti, A. Sequential labelling and DNABERT For splice site prediction in Homo Sapiens DNA. Preprint at https://arXiv.org/quant-ph/2212.07638 (2022).
Zhou, Z. et al. DNABERT-2: efficient foundation model and benchmark for multi-species genome. In Twelfth International Conference on Learning Representations (2024).
Altenburg, T., Giese, S., Wang, S., Muth, T. & Renard, B. Ad hoc learning of peptide fragmentation from mass spectra enables an interpretable detection of phosphorylated and cross-linked peptides. Nat. Mach. Intell. 4, 378–388 (2022).
Raffel, C. et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 21, 5485–5551 (2020).
Liang, S. et al. Rm-LR: a long-range-based deep learning model for predicting multiple types of RNA modifications. Comput. Biol. Med. 164, 107238 (2023).
Chen, J. et al. Interpretable RNA foundation model from unannotated data for highly accurate RNA structure and function predictions. Preprint at bioRxiv https://doi.org/10.1101/2022.08.06.503062 (2022).
Holbrook, S. RNA structure: the long and the short of it. Curr. Opin. Struct. Biol. 15, 302–308 (2005).
Sweeney, B. et al. RNAcentral 2021: secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 49, D212–D220 (2021).
Leontis, N., Lescoute, A. & Westhof, E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 16, 279–287 (2006).
Fierro-Monti, I. & Mathews, M. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci. 25, 241–246 (2000).
Butcher, S. & Pyle, A. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 44, 1302–1311 (2011).
Lee, Y. et al. Surgical fine-tuning improves adaptation to distribution shifts. In Eleventh International Conference on Learning Representations (2023).
Moon, K. et al. Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 37, 1482–1492 (2019).
Kirk, J. et al. Functional classification of long non-coding RNAs by k-mer content. Nat. Genet. 50, 1474–1482 (2018).
Akiyama, M. & Sakakibara, Y. Informative RNA base embedding for RNA structural alignment and clustering by deep representation learning. NAR Genom. Bioinform. 4, lqac012 (2022).
Zhang, Y. et al. Multiple sequence alignment-based RNA language model and its application to structural inference. Nucleic Acids Res. 52, e3–e3 (2024).
Fiannaca, A., La Rosa, M., La Paglia, L., Rizzo, R. & Urso, A. nRC: non-coding RNA classifier based on structural features. BioData Min. 10, 1–18 (2017).
Nawrocki, E. et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43, D130–D137 (2015).
Wen, M., Cong, P., Zhang, Z., Lu, H. & Li, T. DeepMirTar: a deep-learning approach for predicting human miRNA targets. Bioinformatics 34, 3781–3787 (2018).
Pla, A., Zhong, X. & Rayner, S. miRAW: a deep learning-based approach to predict microRNA targets by analyzing whole microRNA transcripts. PLoS Comput. Biol. 14, e1006185 (2018).
Enright, A. et al. MicroRNA targets in Drosophila. Genome Biol. 4, 1–27 (2003).
Krüger, J. & Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451–W454 (2006).
Pita, T., Feliciano, J. & Leitão, J. Identification of Burkholderia cenocepacia non-coding RNAs expressed during Caenorhabditis elegans infection. Appl. Microbiol. Biotechnol. 107, 3653–3671 (2023).
Agarwal, V., Bell, G., Nam, J. & Bartel, D. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015).
Ding, J., Li, X. & Hu, H. TarPmiR: a new approach for microRNA target site prediction. Bioinformatics 32, 2768–2775 (2016).
Fu, L. et al. UFold: fast and accurate RNA secondary structure prediction with deep learning. Nucleic Acids Res. 50, e14–e14 (2022).
Tan, Z., Fu, Y., Sharma, G. & Mathews, D. TurboFold II: RNA structural alignment and secondary structure prediction informed by multiple homologs. Nucleic Acids Res. 45, 11570–11581 (2017).
Sloma, M. & Mathews, D. Exact calculation of loop formation probability identifies folding motifs in RNA secondary structures. RNA 22, 1808–1818 (2016).
Danaee, P. et al. bpRNA: large-scale automated annotation and analysis of RNA secondary structure. Nucleic Acids Res. 46, 5381–5394 (2018).
Sun, Y. et al. Ernie 2.0: a continual pre-training framework for language understanding. In Proc. AAAI Conference on Artificial Intelligence 34 (eds Wooldridge, M., Dy, J. & Natarajan, S.) 8968–8975 (AAAI, 2020).
Vaswani, A. et al. Attention is all you need. In Proc. Advances in Information Processing Systems 30 (eds Guyon, I. et al.) 5999–6009 (NeurIPS, 2017).
Karpicke, J. D., Lehman, M. & Aue, W. R. Retrieval-based learning: an episodic context account. In Psychology of Learning and Motivation Vol. 61, 237–284 (Academic Press, 2014).
Joshi, M. et al. SpanBERT: improving pre-training by representing and predicting spans. Trans. Assoc. Comput. Linguist. 8, 64–77 (2020).
Wu, R. et al. High-resolution de novo structure prediction from primary sequence. Preprint at bioRxiv https://doi.org/10.1101/2022.07.21.500999 (2022).
Giudice, G., Sánchez-Cabo, F., Torroja, C. & Lara-Pezzi, E. ATtRACT—a database of RNA-binding proteins and associated motifs. Database 2016, baw035 (2016).
Piva, F., Giulietti, M., Burini, A. & Principato, G. SpliceAid 2: a database of human splicing factors expression data and RNA target motifs. Hum. Mutat. 33, 81–85 (2012).
Fang, Y., Pan, X. & Shen, H. Recent deep learning methodology development for RNA-RNA interaction prediction. Symmetry 14, 1302 (2022).
Gu, T., Zhao, X., Barbazuk, W. & Lee, J. miTAR: a hybrid deep learning-based approach for predicting miRNA targets. BMC Bioinform. 22, 1–16 (2021).
Sato, K., Akiyama, M. & Sakakibara, Y. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat. Commun. 12, 1–9 (2021).
Zuker, M. & Stiegler, P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 9, 133–148 (1981).
Frankish, A. et al. GENCODE 2021. Nucleic Acids Res. 49, D916–D923 (2021).
Ning, W. CatIIIIIIII/RNAErnie: v.1.0. Zenodo https://doi.org/10.5281/zenodo.10847621 (2024).
Ning, W. RNAErnie docker. Zenodo https://doi.org/10.5281/zenodo.10847856 (2024).
Ning, W. CatIIIIIIII/RNAErnie_baselines: v.1.0.0. Zenodo https://doi.org/10.5281/zenodo.10851577 (2024).
Panwar, B., Arora, A. & Raghava, G. Prediction and classification of ncRNAs using structural information. BMC Genomics 15, 1–13 (2014).
Wang, L. et al. ncRFP: a novel end-to-end method for non-coding RNAs family prediction based on deep learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 18, 784–789 (2020).
Deng, C. et al. RNAGCN: RNA tertiary structure assessment with a graph convolutional network. Chin. Phys. B 31, 118702 (2022).
Chantsalnyam, T., Lim, D., Tayara, H. & Chong, K. ncRDeep: non-coding RNA classification with convolutional neural network. Comput. Biol. Chem. 88, 107364 (2020).
Chantsalnyam, T., Siraj, A., Tayara, H. & Chong, K. ncRDense: a novel computational approach for classification of non-coding RNA family by deep learning. Genomics 113, 3030–3038 (2021).
Reuter, J. & Mathews, D. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinform. 11, 1–9 (2010).
Andronescu, M., Aguirre-Hernandez, R., Condon, A. & Hoos, H. RNAsoft: a suite of RNA secondary structure prediction and design software tools. Nucleic Acids Res. 31, 3416–3422 (2003).
Lorenz, R. et al. ViennaRNA package 2.0. Algorithms Mol. Biol. 6, 1–14 (2011).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Huang, L. et al. LinearFold: linear-time approximate RNA folding by 5′-to-3′ dynamic programming and beam search. Bioinformatics 35, i295–i304 (2019).
Wayment-Steele, H. K. et al. RNA secondary structure packages evaluated and improved by high-throughput experiments. Nat. Methods 19, 1234–1242 (2022).
Chen, X., Li, Y., Umarov, R., Gao, X. & Song, L. RNA secondary structure prediction by learning unrolled algorithms. In International Conference on Learning Representations (2020).
Do, C., Woods, D. & Batzoglou, S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics 22, e90–e98 (2006).
Zakov, S., Goldberg, Y., Elhadad, M. & Ziv-Ukelson, M. Rich parameterization improves RNA structure prediction. J. Comput. Biol. 18, 1525–1542 (2011).
Acknowledgements
This work is kindly supported by the National Science and Technology Major Project under grant no. 2021ZD0110303 (N.W., J.B., X.L. and H.X.) and the National Science Foundation of China under grant no. 62141220 (Y.L. and L.K.).
Author information
Authors and Affiliations
Contributions
All authors made contributions to this paper. N.W. and J.B. conducted experiments and wrote part of the paper. Y.L., X.L. and S.M. were involved in the discussion and wrote part of the paper. L.K. oversaw the research progress, was involved in the discussion and wrote part of the paper. H.X. oversaw the research progress, designed the study and experiments, was involved in the discussion and wrote the paper. H.X. is the senior author, and L.K. is the co-senior contributor.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Xiangfu Zhong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The figure illustrates the use of a special ‘[IND]’ token followed by the RNAcentral instance type as an indicator.
During the pre-training phase, the instance type is masked out and RNAErnie attempts to predict it. In downstream tasks, a two-stage pipeline is employed, which aggregates the top-K predicted indicators to improve performance.
Supplementary information
Supplementary Information
Related work, Supplementary Figs. 1 and 2, and results and analysis.
Source data
Source Data Fig. 1
Scatter coordinates and labels.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, N., Bian, J., Li, Y. et al. Multi-purpose RNA language modelling with motif-aware pretraining and type-guided fine-tuning. Nat Mach Intell 6, 548–557 (2024). https://doi.org/10.1038/s42256-024-00836-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-024-00836-4