Abstract
Deep neural-network-based language models (LMs) are increasingly applied to large-scale protein sequence data to predict protein function. However, being largely black-box models and thus challenging to interpret, current protein LM approaches do not contribute to a fundamental understanding of sequence–function mappings, hindering rule-based biotherapeutic drug development. We argue that guidance drawn from linguistics, a field specialized in analytical rule extraction from natural language data, can aid with building more interpretable protein LMs that are more likely to learn relevant domain-specific rules. Differences between protein sequence data and linguistic sequence data require the integration of more domain-specific knowledge in protein LMs compared with natural language LMs. Here, we provide a linguistics-based roadmap for protein LM pipeline choices with regard to training data, tokenization, token embedding, sequence embedding and model interpretation. Incorporating linguistic ideas into protein LMs enables the development of next-generation interpretable machine learning models with the potential of uncovering the biological mechanisms underlying sequence–function relationships.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




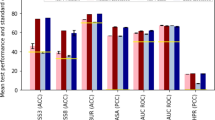
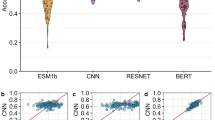
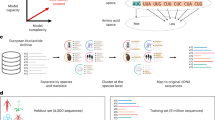
Similar content being viewed by others
References
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conf. of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies Vol. 1, 4171–4186 (Association for Computational Linguistics, 2019).
Liu, Y. et al. RoBERTa: A Robustly Optimized BERT Pretraining Approach. Preprint at https://doi.org/10.48550/arXiv.1907.11692 (2019).
Brown, T. B. et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 33, 1877–1901 (2020).
Bepler, T. & Berger, B. Learning the protein language: evolution, structure, and function. Cell Syst. 12, 654–669 (2021).
Ofer, D., Brandes, N. & Linial, M. The language of proteins: NLP, machine learning & protein sequences. Comput. Struct. Biotechnol. J. 19, 1750–1758 (2021).
Alley, E. C., Khimulya, G., Biswas, S., AlQuraishi, M. & Church, G. M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 16, 1315–1322 (2019).
Brandes, N., Ofer, D., Peleg, Y., Rappoport, N. & Linial, M. ProteinBERT: a universal deep-learning model of protein sequence and function. Bioinformatics 38, 2102–2110 (2022).
Elnaggar, A. et al. ProtTrans: Toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2022).
Heinzinger, M. et al. Modeling aspects of the language of life through transfer-learning protein sequences. BMC Bioinf. 20, 723 (2019).
Rao, R. et al. Evaluating protein transfer learning with TAPE. Adv. Neural Inf. Process. Syst. 32, 9689–9701 (2019).
Hie, B. L., Yang, K. K. & Kim, P. S. Evolutionary velocity with protein language models predicts evolutionary dynamics of diverse proteins. Cell Syst. 13, 274–285 (2022).
Unsal, S. et al. Learning functional properties of proteins with language models. Nat. Mach. Intell. 4, 227–245 (2022).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl Acad. Sci. USA 118, e2016239118 (2021).
Meier, J. et al. Language models enable zero-shot prediction of the effects of mutations on protein function. Adv. Neural Inf. Proc. Syst. 34, 29287–29303 (2021).
Wang, Y. et al. A high efficient biological language model for predicting protein–protein interactions. Cells 8, 122 (2019).
Xu, M. et al. PEER: a comprehensive and multi-task benchmark for protein sequence understanding. In International Conference of Learning Representations (ICLR, 2022).
Nijkamp, E., Ruffolo, J., Weinstein, E. N., Naik, N. & Madani, A. ProGen2: exploring the boundaries of protein language models. Preprint at https://doi.org/10.48550/arXiv.2206.13517 (2022).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Ferruz, N., Schmidt, S. & Höcker, B. ProtGPT2 is a deep unsupervised language model for protein design. Nat. Commun. 13, 4348 (2022).
Sundermeyer, M., Schlüter, R. & Ney, H. LSTM neural networks for language modeling. Interspeech 2012 194–197 https://doi.org/10.21437/Interspeech.2012-65; (ISCA, 2012).
Burley, S. K. et al. in Protein Crystallography: Methods and Protocols (eds. Wlodawer, A., Dauter, Z. & Jaskolski, M.) 627–641 (Springer, 2017).
Olsen, T. H., Boyles, F. & Deane, C. M. Observed Antibody Space: a diverse database of cleaned, annotated, and translated unpaired and paired antibody sequences. Protein Sci. 31, 141–146 (2022).
Corrie, B. D. et al. iReceptor: a platform for querying and analyzing antibody/B-cell and T-cell receptor repertoire data across federated repositories. Immunol. Rev. 284, 24–41 (2018).
Ferruz, N. & Höcker, B. Controllable protein design with language models. Nat. Mach. Intell. 4, 521–532 (2022).
Vig, J. et al. BERTology meets biology: Interpreting attention in protein language models. In International Conference on Learning Representations (ICLR, 2020).
Akbar, R. et al. Progress and challenges for the machine learning-based design of fit-for-purpose monoclonal antibodies. mAbs 14, 2008790 (2022).
Naseem, U., Razzak, I., Khan, S. K. & Prasad, M. A comprehensive survey on word representation models: from classical to state-of-the-art word representation language models. ACM Trans. Asian Low-Resour. Lang. Inf. Process. 20, 1–35 (2021).
Lin, T., Wang, Y., Liu, X. & Qiu, X. A survey of transformers. AI Open 3, 111–132 (2022).
Kaplan, J. et al. Scaling laws for neural language models. Preprint at https://doi.org/10.48550/arXiv.2001.08361 (2020).
Rae, J. W. et al. Scaling language models: methods, analysis & insights from training gopher. Preprint at https://doi.org/10.48550/arXiv.2112.11446 (2022).
Villegas-Morcillo, A. et al. Unsupervised protein embeddings outperform hand-crafted sequence and structure features at predicting molecular function. Bioinformatics 37, 162–170 (2021).
Firth, J. R. In Selected Papers of J. R. Firth 1952–1959 (ed. Palmer, F. R.) 168–205 (Longmans, 1968).
Greiff, V. et al. Systems analysis reveals high genetic and antigen-driven predetermination of antibody repertoires throughout B cell development. Cell Rep. 19, 1467–1478 (2017).
Elhanati, Y. et al. Inferring processes underlying B-cell repertoire diversity. Philos. Trans. R. Soc. B 370, 20140243 (2015).
Kutuzov, A. & Kuzmenko, E. To lemmatize or not to lemmatize: how word normalisation affects ELMo performance in word sense disambiguation. In Proc. First NLPL Workshop on Deep Learning for Natural Language Processing 22–28 (Linköping University Electronic Press, 2019).
Pan, Y., Li, X., Yang, Y. & Dong, R. Morphological word segmentation on agglutinative languages for neural machine translation. Preprint at https://doi.org/10.48550/arXiv.2001.01589 (2020).
Schwartz, L. et al. Neural polysynthetic language modelling. Preprint at https://doi.org/10.48550/arXiv.2005.05477 (2020).
Szklarczyk, D. et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Rogers, A., Kovaleva, O. & Rumshisky, A. A primer in BERTology: what we know about how BERT works. Trans. Assoc. Comput. Linguist. 8, 842–866 (2021).
Mielke, S. J. et al. Between words and characters: a brief history of open-vocabulary modeling and Tokenization in NLP. Preprint at https://doi.org/10.48550/arXiv.2112.10508 (2021).
Lee, J. et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36, 1234–1240 (2020).
Qiu, X. et al. Pre-trained models for natural language processing: a survey. Sci. China Technol. Sci. 63, 1872–1897 (2020).
Bender, E. M., Gebru, T., McMillan-Major, A. & Shmitchell, S. On the dangers of stochastic parrots: can language models be too big? In Proc. 2021 ACM Conference on Fairness, Accountability, and Transparency 610–623 (Association for Computing Machinery, 2021).
Doddapaneni, S., Ramesh, G., Khapra, M. M., Kunchukuttan, A. & Kumar, P. A primer on pretrained multilingual language models. Preprint at https://doi.org/10.48550/arXiv.2107.00676 (2021).
Shin, S. et al. On the effect of pretraining corpora on in-context Learning by a large-scale language model. In Proc. 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 5168–5186 (Association for Computational Linguistics, 2022).
Leem, J., Mitchell, L. S., Farmery, J. H. R., Barton, J. & Galson, J. D. Deciphering the language of antibodies using self-supervised learning. Patterns 3, 100513 (2022).
Ruffolo, J. A., Gray, J. J. & Sulam, J. Deciphering antibody affinity maturation with language models and weakly supervised learning. In Machine Learning in Structural Biology Workshop, NeurIPS (2021)
Olsen, T. H., Moal, I. H. & Deane, C. M. AbLang: an antibody language model for completing antibody sequences. Bioinformatics Advances 2, vbac046 (2022).
Conneau, A. et al. Unsupervised Cross-lingual Representation Learning at Scale. In Proc. 58th Annual Meeting of the Association for Computational Linguistics 8440–8451 (Association for Computational Linguistics, 2020).
Agerri, R. et al. Give your Text Representation Models some Love: the Case for Basque. in Proceedings of the Twelfth Language Resources and Evaluation Conference 4781–4788 (European Language Resources Association, 2020).
Liu, C.-L., Hsu, T.-Y., Chuang, Y.-S. & Lee, H.-Y. A study of cross-lingual ability and language-specific information in multilingual BERT. Preprint at https://doi.org/10.48550/arXiv.2004.09205 (2020).
Lauscher, A., Ravishankar, V., Vulić, I. & Glavaš, G. From zero to hero: on the limitations of zero-shot language transfer with multilingual transformers. In Proc. 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP) 4483–4499 (Association for Computational Linguistics, 2020).
de Vries, W., Wieling, M. & Nissim, M. Make the Best of Cross-lingual Transfer: Evidence from POS Tagging with over 100 Languages. In Proc. 60th Annual Meeting of the Association for Computational Linguistics (2022).
Ruffolo, J. A., Sulam, J. & Gray, J. J. Antibody structure prediction using interpretable deep learning. Patterns 3, 100406 (2022).
Shuai, R. W., Ruffolo, J. A. & Gray, J. J. Generative language modeling for antibody design. In Machine Learning for Structural Biology Workshop, NeurIPS (2021)
Ostrovsky-Berman, M., Frankel, B., Polak, P. & Yaari, G. Immune2vec: embedding B/T cell receptor sequences in ℝN using natural language processing. Front. Immunol. 12, 680687 (2021).
Kao, W.-T. & Lee, H. Is BERT a cross-disciplinary knowledge learner? A surprising finding of pre-trained models’ transferability. In Findings of the Association for Computational Linguistics: EMNLP 2021 2195–2208 (Association for Computational Linguistics, 2021).
Krishna, K., Bigham, J. & Lipton, Z. C. Does pretraining for summarization require knowledge transfer? In Findings of the Association for Computational Linguistics: EMNLP 2021 3178–3189 (Association for Computational Linguistics, 2021).
Robert, P. A. et al. Unconstrained generation of synthetic antibody–antigen structures to guide machine learning methodology for antibody specificity prediction. Nat. Comput. Sci. 2, 845–865 (2022).
Marcou, Q., Mora, T. & Walczak, A. M. High-throughput immune repertoire analysis with IGoR. Nat. Commun. 9, 561 (2018).
Weber, C. R. et al. immuneSIM: tunable multi-feature simulation of B- and T-cell receptor repertoires for immunoinformatics benchmarking. Bioinformatics 36, 3594–3596 (2020).
Morris, T. P., White, I. R. & Crowther, M. J. Using simulation studies to evaluate statistical methods. Stat. Med. 38, 2074–2102 (2019).
Yang, K. K., Wu, Z., Bedbrook, C. N. & Arnold, F. H. Learned protein embeddings for machine learning. Bioinformatics 34, 2642–2648 (2018).
Brown, P. F., Pietra, V. J. D., Mercer, R. L., Pietra, S. A. D. & Lai, J. C. An estimate of an upper bound for the entropy of English. Comput. Linguist. 18, 10 (1992).
Xu, J., Zhou, H., Gan, C., Zheng, Z. & Li, L. Vocabulary Learning via Optimal Transport for Neural Machine Translation. In Proc. 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing Vol. 1: Long Papers, 7361–7373 (Association for Computational Linguistics, 2021).
Gage, P. A new algorithm for data compression. C Users J. 12, 23–38 (1994).
Pinter, Y. Integrating Approaches to Word Representation. ArXiv210904876 Cs (2021).
Hofmann, V., Pierrehumbert, J. & Schütze, H. Superbizarre is not superb: derivational morphology improves BERT’s interpretation of complex words. In Proc. 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing Vol. 1, 3594–3608 (Association for Computational Linguistics, 2021).
Littmann, M., Heinzinger, M., Dallago, C., Olenyi, T. & Rost, B. Embeddings from deep learning transfer GO annotations beyond homology. Sci. Rep. 11, 1160 (2021).
Madani, A. et al. Deep neural language modeling enables functional protein generation across families. Preprint at bioRxiv https://doi.org/10.1101/2021.07.18.452833 (2021).
Devi, G., Tendulkar, A. V. & Chakraborti, S. Protein word detection using text segmentation techniques. In BioNLP 2017 238–246 (Association for Computational Linguistics, 2017).
Asgari, E., McHardy, A. C. & Mofrad, M. R. K. Probabilistic variable-length segmentation of protein sequences for discriminative motif discovery (DiMotif) and sequence embedding (ProtVecX). Sci. Rep. 9, 3577 (2019).
Szymborski, J. & Emad, A. RAPPPID: towards generalizable protein interaction prediction with AWD-LSTM twin networks. Bioinformatics 38, 3958–3967 (2022).
Strait, B. J. & Dewey, T. G. The Shannon information entropy of protein sequences. Biophys. J. 71, 148–155 (1996).
Shannon, C. E. Prediction and entropy of printed English. Bell Syst. Tech. J. 30, 50–64 (1951).
Asgari, E. & Mofrad, M. R. K. Continuous distributed representation of biological sequences for deep proteomics and genomics. PLoS ONE 10, e0141287 (2015).
Hofmann, V., Schütze, H. & Pierrehumbert, J. An embarrassingly simple method to mitigate undesirable properties of pretrained language model tokenizers. in Proc. 60th Annual Meeting of the Association for Computational Linguistics Vol. 2: Short Papers, 385–393 (Association for Computational Linguistics, 2022).
Matthews, A., Neubig, G. & Dyer, C. Using morphological knowledge in open-vocabulary neural language models. In Proc. 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies Vol. 1: Long Papers, 1435–1445 (Association for Computational Linguistics, 2018).
Gutierrez-Vasques, X., Bentz, C., Sozinova, O. & Samardzic, T. From characters to words: the turning point of BPE merges. In Proc. 16th Conference of the European Chapter of the Association for Computational Linguistics Main Vol., 3454–3468 (Association for Computational Linguistics, 2021).
Alva, V., Söding, J. & Lupas, A. N. A vocabulary of ancient peptides at the origin of folded proteins. eLife 4, e09410 (2015).
Kolodny, R., Nepomnyachiy, S., Tawfik, D. S. & Ben-Tal, N. Bridging themes: short protein segments found in different architectures. Mol. Biol. Evol. 38, 2191–2208 (2021).
Fernandez-Fuentes, N., Dybas, J. M. & Fiser, A. Structural characteristics of novel protein folds. PLoS Comput. Biol. 6, e1000750 (2010).
Ferruz, N. et al. Identification and analysis of natural building blocks for evolution-guided fragment-based protein design. J. Mol. Biol. 432, 3898–3914 (2020).
Akbar, R. et al. A compact vocabulary of paratope-epitope interactions enables predictability of antibody-antigen binding. Cell Rep. 34, 108856 (2021).
Hiraoka, T., Takase, S., Uchiumi, K., Keyaki, A. & Okazaki, N. Optimizing Word Segmentation for Downstream Task. In Findings of the Association for Computational Linguistics: EMNLP 2020 1341–1351 (Association for Computational Linguistics, 2020).
Welleck, S., Brantley, K., Daumé, H. III & Cho, K. Non-monotonic sequential text generation. In Proc. 36th International Conference on Machine Learning 6716–6726 (PMLR, 2019).
Stern, M., Chan, W., Kiros, J. & Uszkoreit, J. Insertion transformer: flexible sequence generation via insertion operations. In Proc. 36th International Conference on Machine Learning 5976–5985 (PMLR, 2019).
Gimona, M. Protein linguistics — a grammar for modular protein assembly? Nat. Rev. Mol. Cell Biol. 7, 68–73 (2006).
Mikolov, T., Yih, W. & Zweig, G. Linguistic regularities in continuous space word representations. In Proc. 2013 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 746–751 (Association for Computational Linguistics, 2013).
Schluter, N. The word analogy testing caveat. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies Vol. 2: Short Papers, 242–246 (Association for Computational Linguistics, 2018).
Hie, B., Zhong, E. D., Berger, B. & Bryson, B. Learning the language of viral evolution and escape. Science 371, 284–288 (2021).
Mikolov, T., Sutskever, I., Chen, K., Corrado, G. S. & Dean, J. Distributed representations of words and phrases and their compositionality. Adv. Neural Inf. Process. Syst. 26 (Curran Associates, Inc., 2013).
Peters, M. E. et al. Deep contextualized word representations. In Proc. 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 1, 2227–2237 (Association for Computational Linguistics, 2018).
Chen, C., Zhou, J., Wang, F., Liu, X. & Dou, D. Structure-aware protein self-supervised learning. Preprint at https://doi.org/10.48550/arXiv.2204.04213 (2022).
Kaufman, S., Rosset, S., Perlich, C. & Stitelman, O. Leakage in data mining: formulation, detection, and avoidance. ACM Trans. Knowl. Discov. Data 6, 15:1–15:21 (2012).
Detlefsen, N. S., Hauberg, S. & Boomsma, W. Learning meaningful representations of protein sequences. Nat. Commun. 13, 1914 (2022).
Montague, R. Universal grammar. Theoria 36, 373–393 (1970).
McCoy, R. T., Frank, R. & Linzen, T. Does syntax need to grow on trees? Sources of hierarchical inductive bias in sequence-to-sequence networks. Trans. Assoc. Comput. Linguist. 8, 125–140 (2020).
Tai, K. S., Socher, R. & Manning, C. D. Improved semantic representations from tree-structured long short-term memory networks. In Proc. 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing Vol. 1: Long Papers, 1556–1566 (Association for Computational Linguistics, 2015).
Linzen, T. What can linguistics and deep learning contribute to each other? Response to Pater. Language 95, e99–e108 (2019).
Ettinger, A. What BERT is not: lessons from a new suite of psycholinguistic diagnostics for language models. Trans. Assoc. Comput. Linguist. 8, 34–48 (2020).
Hu, J., Gauthier, J., Qian, P., Wilcox, E. & Levy, R. A systematic assessment of syntactic generalization in neural language models. In Proc. 58th Annual Meeting of the Association for Computational Linguistics 1725–1744 (Association for Computational Linguistics, 2020).
McCoy, R. T., Pavlick, E. & Linzen, T. Right for the wrong reasons: diagnosing syntactic heuristics in natural language inference. In Proc. 57th Annual Meeting of the Association for Computational Linguistics 3428–3448 (Association for Computational Linguistics, 2019).
McCoy, R. T., Smolensky, P., Linzen, T., Gao, J. & Celikyilmaz, A. How much do language models copy from their training data? Evaluating linguistic novelty in text generation using RAVEN. Preprint at https://doi.org/10.48550/arXiv.2111.09509 (2021).
Niven, T. & Kao, H.-Y. Probing Neural Network Comprehension of Natural Language Arguments. in Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics 4658–4664 (Association for Computational Linguistics, 2019). https://doi.org/10.18653/v1/P19-1459
Clark, K., Khandelwal, U., Levy, O. & Manning, C. D. What does BERT look at? An analysis of BERT’s attention. In Proc. 2019 ACL Workshop BlackboxNLP: Analyzing and Interpreting Neural Networks for NLP 276–286 (Association for Computational Linguistics, 2019).
Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 1, 206–215 (2019).
Adebayo, J., Muelly, M., Abelson, H. & Kim, B. Post hoc explanations may be ineffective for detecting unknown spurious correlation. In International Conference on Learning Representations (2022).
Linzen, T., Dupoux, E. & Goldberg, Y. Assessing the ability of LSTMS to learn syntax-sensitive dependencies. Trans. Assoc. Comput. Linguist. 4, 521–535 (2016).
Goldberg, Y. Assessing BERT’s syntactic abilities. Preprint at https://doi.org/10.48550/arXiv.1901.05287 (2019).
Warstadt, A. et al. BLiMP: a benchmark of linguistic minimal pairs for English. Proc. Soc. Comput. Linguist. 3, 437–438 (2020).
Gold, E. M. Language identification in the limit. Inf. Control 10, 447–474 (1967).
Angluin, D. Learning regular sets from queries and counterexamples. Inf. Comput. 75, 87–106 (1987).
Weiss, G., Goldberg, Y. & Yahav, E. Extracting automata from recurrent neural networks using queries and counterexamples. In Proc. 35th International Conference on Machine Learning 5247–5256 (PMLR, 2018).
Angluin, D. Computational learning theory: survey and selected bibliography. In Proc. 24th Annual ACM Symposium on Theory of Computing 351–369 (Association for Computing Machinery, 1992).
Eyraud, R. & Ayache, S. Distillation of weighted automata from recurrent neural networks using a spectral approach. Mach. Learn. https://doi.org/10.1007/s10994-021-05948-1 (2021).
Wang, Q. et al. An empirical evaluation of rule extraction from recurrent neural networks. Neural Comput. 30, 2568–2591 (2018).
Sandve, G. K. & Greiff, V. Access to ground truth at unconstrained size makes simulated data as indispensable as experimental data for bioinformatics methods development and benchmarking. Bioinformatics 38, 4994–4996 (2022).
Tenney, I., Das, D. & Pavlick, E. BERT rediscovers the classical NLP pipeline. In Proc. 57th Annual Meeting of the Association for Computational Linguistics 4593–4601 (Association for Computational Linguistics, 2019).
Bhattamishra, S., Ahuja, K. & Goyal, N. On the ability and limitations of transformers to recognize formal languages. In Proc. 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP) 7096–7116 (Association for Computational Linguistics, 2020).
Clark, P., Tafjord, O. & Richardson, K. Transformers as soft reasoners over language. In Proc. 29th International Joint Conference on Artificial Intelligence Vol. 4 3882–3890 (2020).
Acknowledgements
We thank K. Cho and E. M. Bender for their comments on the manuscript. We acknowledge support by the Leona M. and Harry B. Helmsley Charitable Trust (2019PG-T1D011, to V.G.), UiO World-Leading Research Community (to V.G.), UiO:LifeScience Convergence Environment Immunolingo (to V.G., G.K.S. and D.T.T.H.), EU Horizon 2020 iReceptorplus (825821 to V.G.), a Research Council of Norway FRIPRO project (300740 to V.G.), a Research Council of Norway IKTPLUSS project (311341 to V.G. and G.K.S.), a Norwegian Cancer Society Grant (215817 to V.G.) and Stiftelsen Kristian Gerhard Jebsen (KG Jebsen Coeliac Disease Research Centre to G.K.S.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
V.G. declares advisory board positions in aiNET GmbH, Enpicom B.V, Specifica Inc, Adaptyv Biosystems, EVQLV and Omniscope. V.G. is a consultant for Roche/Genentech, immunai and Proteinea. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Tunca Dogan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vu, M.H., Akbar, R., Robert, P.A. et al. Linguistically inspired roadmap for building biologically reliable protein language models. Nat Mach Intell 5, 485–496 (2023). https://doi.org/10.1038/s42256-023-00637-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-023-00637-1
This article is cited by
-
DisoFLAG: accurate prediction of protein intrinsic disorder and its functions using graph-based interaction protein language model
BMC Biology (2024)
-
Adaptive immune receptor repertoire analysis
Nature Reviews Methods Primers (2024)
-
AI protein shake-up
Nature Machine Intelligence (2024)
-
Language models and linguistic theories beyond words
Nature Machine Intelligence (2023)