Abstract
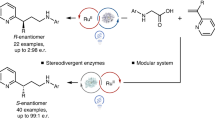
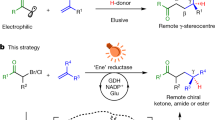
State-of-the-art non-natural photoenzymatic catalysis typically relies on either ultraviolet light activation or the formation of electron donor–acceptor complexes. Inspired by nature, herein we develop a biocatalytic scheme based on direct visible-light excitation of flavoprotein to achieve stereocontrolled intermolecular radical hydroarylation of alkenes with electron-rich arenes. Following visible-light activation, flavin-dependent ene-reductases—which naturally catalyse the two-electron reduction of activated alkenes—are repurposed to trigger single-electron oxidation of arenes and afford C(sp2)–C(sp3) bond-forming products in a redox-neutral and enantiodivergent fashion while both enantiomers are obtained with different enzymes. Experiments and computational simulations demonstrate that the reaction is initiated from the single-electron oxidation of substrate by the direct visible-light-excited flavoprotein, and explain the origin of enantioselectivity in this radical hydroarylation that is otherwise notoriously challenging to achieve by chemocatalysis. This work expands the repertoire of photoenzymatic catalysis and will stimulate the development of further biotransformations.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data relating to the materials and methods, experimental procedures, mechanistic studies and computational calculations, HPLC spectra and NMR spectra are available in the Supplementary Information or from the authors on reasonable request. The atomic coordinates of the optimized computational models are available in the Supplementary Data. The configurations of molecular dynamics simulations have been deposited in GitHub (https://github.com/fpikachu96/OYE1-GluER). Crystallographic data for the structures reported in this article have been deposited at the Cambridge Crystallographic Data Centre under deposition no. CCDC 2226701 (3aa). Copies of the data can be obtained free of charge via https://www.ccdc.cam.ac.uk/structures/.
References
Chen, K. & Arnold, F. H. Engineering new catalytic activities in enzymes. Nat. Catal. 3, 203–213 (2020).
Hughes, D. L. Highlights of the recent patent literature-focus on biocatalysis innovation. Org. Process Res. Dev. 26, 1878–1899 (2022).
Winkler, C. K., Schrittwieser, J. H. & Kroutil, W. Power of biocatalysis for organic synthesis. ACS Cent. Sci. 7, 55–71 (2021).
Leveson-Gower, R. B., Mayer, C. & Roelfes, G. The importance of catalytic promiscuity for enzyme design and evolution. Nat. Rev. Chem. 3, 687–705 (2019).
Reetz, M. Making enzymes suitable for organic chemistry by rational protein design. ChemBioChem 23, e202200049 (2022).
Yang, Y. & Arnold, F. H. Navigating the unnatural reaction space: directed evolution of heme proteins for selective carbene and nitrene transfer. Acc. Chem. Res. 54, 1209–1225 (2021).
Fasan, R. et al. Biocatalytic strategy for construction of sp3-rich polycyclic compounds from directed evolution and computational modeling. Preprint available at https://doi.org/10.21203/rs.3.rs-1639676/v1
Rui, J. et al. Directed evolution of nonheme iron enzymes to access abiological radical-relay C(sp3)−H azidation. Science 376, 869–874 (2022).
Zhou, Q., Chin, M., Fu, Y., Liu, P. & Yang, Y. Stereodivergent atom-transfer radical cyclization by engineered cytochromes P450. Science 374, 1612–1616 (2021).
Liang, A. D., Serrano-Plana, J., Peterson, R. L. & Ward, T. R. Artificial metalloenzymes based on the biotin-streptavidin technology: enzymatic cascades and directed evolution. Acc. Chem. Res. 52, 585–595 (2019).
Huang, J. et al. Unnatural biosynthesis by an engineered microorganism with heterologously expressed natural enzymes and an artificial metalloenzyme. Nat. Chem. 13, 1186–1191 (2021).
Stephenson, C. R. J., Yoon, T. P. & MacMillan, D. W. C. Visible Light Photocatalysis in Organic Chemistry (Wiley-VCH, 2018).
Harrison, W., Huang, X. & Zhao, H. Photobiocatalysis for abiological transformations. Acc. Chem. Res. 55, 1087–1096 (2022).
Sorigue, D. et al. An algal photoenzyme converts fatty acids to hydrocarbons. Science 357, 903–907 (2017).
Sorigue, D. et al. Mechanism and dynamics of fatty acid photodecarboxylase. Science 372, eabd5687 (2021).
Amer, M. et al. Low carbon strategies for sustainable bio-alkane gas production and renewable energy. Energy Environ. Sci. 13, 1818–1831 (2020).
Litman, Z. C., Wang, Y., Zhao, H. & Hartwig, J. F. Cooperative asymmetric reactions combining photocatalysis and enzymatic catalysis. Nature 560, 355–359 (2018).
Ye, Y. et al. Using enzymes to tame nitrogen-centred radicals for enantioselective hydroamination. Nat. Chem. 15, 206–212 (2023).
Liu, X. et al. A genetically encoded photosensitizer protein facilitates the rational design of a miniature photocatalytic CO2-reducing enzyme. Nat. Chem. 10, 1201–1206 (2018).
Sun, N. et al. Enantioselective [2 + 2]-cycloadditions with triplet photoenzymes. Nature 611, 715–720 (2022).
Trimble, J. S. et al. A designed photoenzyme for enantioselective [2+2] cycloadditions. Nature 611, 709–714 (2022).
Crisenza, G. E. M., Mazzarella, D. & Melchiorre, P. Synthetic methods driven by the photoactivity of electron donor-acceptor complexes. J. Am. Chem. Soc. 142, 5461–5476 (2020).
Emmanuel, M. A., Greenberg, N. R., Oblinsky, D. G. & Hyster, T. K. Accessing non-natural reactivity by irradiating nicotinamide-dependent enzymes with light. Nature 540, 414–417 (2016).
Fu, H. et al. An asymmetric sp3–sp3 cross-electrophile coupling using ‘ene’-reductases. Nature 610, 302–307 (2022).
Biegasiewicz, K. F. et al. Photoexcitation of flavoenzymes enables a stereoselective radical cyclization. Science 364, 1166–1169 (2019).
Huang, X. et al. Photoenzymatic enantioselective intermolecular radical hydroalkylation. Nature 584, 69–74 (2020).
Huang, X. et al. Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition. Nat. Catal. 5, 586–593 (2022).
Peng, Y. et al. Photoinduced promiscuity of cyclohexanone monooxygenase for the enantioselective synthesis of α-fluoroketones. Angew. Chem. Int. Ed. 61, e202211199 (2022).
Duan, X. et al. A photoenzymatic strategy for radical-mediated stereoselective hydroalkylation with diazo compounds. Angew. Chem. Int. Ed. 62, e202214135 (2023).
De Angelis, M., Iazzetti, A., Serraiocco, A. & Ciogli, A. Asymmetric hydroarylation reactions catalyzed by transition metals: last 10 years in a mini review. Catalysts 12, 1289 (2022).
Zhang, P., Tsuji, N., Ouyang, J. & List, B. Strong and confined acids catalyze asymmetric intramolecular hydroarylations of unactivated olefins with indoles. J. Am. Chem. Soc. 143, 675–680 (2021).
Chen, B. H., Du, Y. D. & Shu, W. Organophotocatalytic regioselective C-H alkylation of electron-rich arenes using activated and unactivated alkenes. Angew. Chem. Int. Ed. 61, e202200773 (2022).
Mondal, S. et al. Enantioselective radical reactions using chiral catalysts. Chem. Rev. 122, 5842–5976 (2022).
Massey, V., Stankovich, M. & Hemmerich, P. Light-mediated reduction of flavoproteins with flavins as catalysts. Biochemistry 17, 1–8 (1978).
Zhuang, B., Ramodiharilafy, R., Liebl, U., Aleksandrov, A. & Vos, M. H. Ultrafast photooxidation of protein-bound anionic flavin radicals. Proc. Natl Acad. Sci. USA 119, e2118924119 (2022).
Toogood, H. S. & Scrutton, N. S. Discovery, characterisation, engineering and applications of ene-reductases for industrial biocatalysis. ACS Catal. 8, 3532–3549 (2018).
Kumar Roy, T., Sreedharan, R., Ghosh, P., Gandhi, T. & Maiti, D. Ene-reductase: a multifaceted biocatalyst in organic synthesis. Chemistry 28, e202103949 (2022).
Stewart, R. C. & Massey, V. Potentiometric studies of native and flavin-substituted old yellow enzyme. J. Biol. Chem. 260, 13639–13647 (1985).
Chen, H.-C. & Swenson, R. P. Effect of the insertion of a glycine residue into the loop spanning residues 536−541 on the semiquinone state and redox properties of the flavin mononucleotide-binding domain of flavocytochrome P450BM-3 from Bacillus megaterium. Biochemistry 47, 13788–13799 (2008).
Case, D. A. et al. AMBER 2018 (Univ. of California, 2018).
Metz, S., Kästner, J., Sokol, A. A., Keal, T. W. & Sherwood, P. ChemShell—a modular software package for QM/MM simulations. WIREs Comput. Mol. Sci. 4, 101–110 (2014).
Ahlrichs, R., Bär, M., Häser, M., Horn, H. & Kölmel, C. Electronic structure calculations on workstation computers: the program system turbomole. Chem. Phys. Lett. 162, 165–169 (1989).
Grimme, S., Ehrlich, S. & Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 32, 1456–1465 (2011).
Acknowledgements
We thank N. Jiao (Peking University), E. Meggers (Philipps University Marburg) and H. Zhao (UIUC) for insightful discussions. This work was supported by the National Key Research and Development Program of China (nos. 2022YFA0913000 to X.H., 2019YFA0405600 to L.Y. and 2019YFA0706900 to C.T.), the National Natural Science Foundation of China (nos. 22277053 to X.H., 22121001 to B.W. and 21927814 and 21825703 to C.T.), the Natural Science Foundation of Jiangsu Province (no. BK20220760 to X.H.) and the Youth Innovation Promotion Association CAS (no. 2022455 to L.Y.). A portion of this work was performed at Steady High Magnetic Field Facilities, High Magnetic Field Laboratory, CAS.
Author information
Authors and Affiliations
Contributions
B.Z. developed the reactions and performed the majority of synthetic experiments. Z.X., B.C. and F.S. assisted with synthetic experiments and mechanistic investigations. F.L. created mutations. J.F. and B.W. conducted computational studies. A.L. and L.Y. carried out EPR measurements and analysis under the supervision of C.T. Y.Z. performed X-ray crystal structure analysis. X.H., B.W. and C.T. wrote the paper with input from all authors. X.H. coordinated and conceived the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Catalysis thanks Jian Xu, Kai Chen, Wei Shu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–37, Tables 1–17 and Methods.
Supplementary Data 1
Atomic coordinates of the optimized computational models.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, B., Feng, J., Yu, L. et al. Direct visible-light-excited flavoproteins for redox-neutral asymmetric radical hydroarylation. Nat Catal 6, 996–1004 (2023). https://doi.org/10.1038/s41929-023-01024-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-023-01024-0