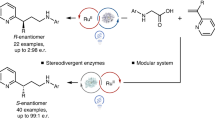
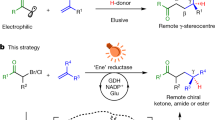
Abstract
Exploiting nature’s catalysts for non-natural transformations that are inaccessible to chemocatalysis is highly desirable but challenging. On the one hand, the widespread nicotinamide-dependent oxidoreductases have not been utilized for single-electron-transfer-induced bimolecular cross-couplings; on the other, the addition of catalytic asymmetric radical conjugate to terminal alkenes remains a challenge owing to strong racemic background reaction and unselective termination of prochiral radical species. Here we report a chemomimetic biocatalysitic approach for construction of alpha-carbonyl stereocentres via an unnatural intermolecular conjugate addition of N-(acyloxy)phthalimides-derived radicals with acceptor-substituted terminal alkenes, by combination of visible-light excitation and nicotinamide-dependent ketoreductases (KREDs). Based on protein crystal structure, we engineered KREDs via a semi-rational mutagenesis strategy to improve reaction outcomes with a small and high-quality variants library. Mechanistic investigations combining wet experiments, crystallographic studies and computational simulations demonstrate that the repurposed biocatalyst can suppress racemic background reaction and unselected side reactions, yielding enantioselectivity that is challenging to achieve by chemocatalysis.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data relating to the materials and methods, experimental procedures, mechanistic studies and computational calculations, HPLC spectra and NMR spectra are available in the Supplementary Information. The atomic coordinates of the optimized computational models are available in the Supplementary Data. The configurations of molecular dynamics simulations have been deposited in GitHub (https://github.com/fpikachu96/KRED-data). The atomic coordinates of apo-P2-D12, apo-K3, P2-D12-2a, K3-2a and K3-NHPI have been deposited in the Protein Data Bank (http://www.rcsb.org) under accession code 7VDO, 7VE7, 7EJJ, 7EJI and 7EJH, respectively. All other data are available from the authors upon reasonable request.
References
Devine, P. N. et al. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2, 409–421 (2018).
Hollmann, F. & Fernandez‐Lafuente, R. Grand challenges in biocatalysis. Front. Catal. 1 https://doi.org/10.3389/fctls.2021.633893 (2021).
Chen, K. & Arnold, F. H. Engineering new catalytic activities in enzymes. Nat. Catal. 3, 203–213 (2020).
Schmermund, L. et al. Photo-biocatalysis: biotransformations in the presence of light. ACS Catal. 9, 4115–4144 (2019).
Emmanuel, M. A., Greenberg, N. R., Oblinsky, D. G. & Hyster, T. K. Accessing non-natural reactivity by irradiating nicotinamide-dependent enzymes with light. Nature 540, 414–417 (2016).
Biegasiewicz, K. F., Cooper, S. J., Emmanuel, M. A., Miller, D. C. & Hyster, T. K. Catalytic promiscuity enabled by photoredox catalysis in nicotinamide-dependent oxidoreductases. Nat. Chem. 10, 770–775 (2018).
Litman, Z. C., Wang, Y., Zhao, H. & Hartwig, J. F. Cooperative asymmetric reactions combining photocatalysis and enzymatic catalysis. Nature 560, 355–359 (2018).
Xu, J. et al. Light-driven kinetic resolution of α-functionalized carboxylic acids enabled by an engineered fatty acid photodecarboxylase. Angew. Chem. Int. Ed. Engl. 58, 8474–8478 (2019).
Biegasiewicz, K. F. et al. Photoexcitation of flavoenzymes enables a stereoselective radical cyclization. Science 364, 1166–1169 (2019).
Black, M. J. et al. Asymmetric redox-neutral radical cyclization catalysed by flavindependent ‘ene’-reductases. Nat. Chem. 12, 71–75 (2020).
Clayman, P. D. & Hyster, T. K. Photoenzymatic generation of unstabilized alkyl radicals: an asymmetric reductive cyclization. J. Am. Chem. Soc. 142, 15673–15677 (2020).
Huang, X. et al. Photoenzymatic enantioselective intermolecular radical hydroalkylation. Nature 584, 69–74 (2020).
Page, C. G. et al. Quaternary charge-transfer complex enables photoenzymatic intermolecular hydroalkylation of olefins. J. Am. Chem. Soc. 143, 97–102 (2021).
Sellés Vidal, L., Kelly, C. L., Mordaka, P. M. & Heap, J. T. Review of NAD(P)H-dependent oxidoreductases: properties, engineering and application. Biochim. Biophys. Acta Proteins Proteom. 1866, 327–347 (2018).
Zheng, C. & You, S.-L. Transfer hydrogenation with Hantzsch esters and related organic hydride donors. Chem. Soc. Rev. 41, 2498–2518 (2012).
Wang, P.-Z., Chen, J.-R. & Xiao, W.-J. Hantzsch esters: an emerging versatile class of reagents in photoredox catalyzed organic synthesis. Org. Biomol. Chem. 17, 6936–6951 (2019).
Kanegusuku, A. L. G. & Roizen, J. L. Recent advances in photoredox-mediated radical conjugate addition reactions: an expanding toolkit for the Giese reaction. Angew. Chem. Int. Ed. Engl. 60, 21116–21149 (2021).
Okada, K., Okamoto, K., Morita, N., Okubo, K. & Oda, M. Photosensitized decarboxylative michael addition through N-(acyloxy)phthalimides via an electron-transfer mechanism. J. Am. Chem. Soc. 113, 9401–9402 (1991).
Zheng, C., Wang, G.-Z. & Shang, R. Catalyst-free decarboxylation and decarboxylative Giese additions of alkyl carboxylates through photoactivation of electron donor-acceptor complex. Adv. Synth. Catal. 361, 4500–4505 (2019).
Chowdhury, R. et al. Decarboxylative alkyl coupling promoted by NADH and blue light. J. Am. Chem. Soc. 142, 20143–20151 (2020).
Crisenza, G. E. M., Mazzarella, D. & Melchiorre, P. Synthetic methods driven by the photoactivity of electron donor–acceptor complexes. J. Am. Chem. Soc. 142, 5461–5476 (2020).
Zhang, J., Li, Y., Xu, R. & Chen, Y. Donor–acceptor complex enables alkoxyl radical generation for metal-free C(sp3)–C(sp3) cleavage and allylation/alkenylation. Angew. Chem. Int. Ed. Engl. 56, 12619–12623 (2017).
Wang, J. et al. Kinetically guided radical-based synthesis of C(sp3)-C(sp3) linkages on DNA. Proc. Natl Acad. Sci. USA 115, E6404–E6410 (2018).
Prier, C. K. & Arnold, F. H. Chemomimetic biocatalysis: exploiting the synthetic potential of cofactor-dependent enzymes to create new catalysts. J. Am. Chem. Soc. 137, 13992–14006 (2015).
Huo, H., Harms, K. & Meggers, E. Catalytic, enantioselective addition of alkyl radicals to alkenes via visible-light-activated photoredox catalysis with a chiral rhodium complex. J. Am. Chem. Soc. 138, 6936–6939 (2016).
Sibi, M. P. & Sausker, J. B. The role of the achiral template in enantioselective transformations. Radical conjugate additions to α-methacrylates followed by hydrogen atom transfer. J. Am. Chem. Soc. 124, 984–991 (2002).
Sibi, M. P. & Patil, K. Enantioselective H-atom transfer reaction: a strategy to synthesize formaldehyde aldol products. Org. Lett. 7, 1453–1456 (2005).
Yin, Y. et al. Conjugate addition–enantioselective protonation of N-aryl glycines to αbranched 2-vinylazaarenes via cooperative photoredox and asymmetric catalysis. J. Am. Chem. Soc. 140, 6083–6087 (2018).
Dai, Z.-Y., Nong, Z.-S. & Wang, P.-S. Light-mediated asymmetric aliphatic C–H alkylation with hydrogen atom transfer catalyst and chiral phosphoric acid. ACS Catal. 10, 4786–4790 (2020).
Rigotti, T. & Alemán, J. Visible light photocatalysis – from racemic to asymmetric activation strategies. Chem. Commun. 56, 11169–11190 (2020).
Zhu, S. & Buchwald, S. L. Enantioselective CuH-catalyzed anti-Markovnikov hydroamination of 1,1-disubstituted alkenes. J. Am. Chem. Soc. 136, 15913–15916 (2014).
Rudroff, F. et al. Opportunities and challenges for combining chemo- and biocatalysis. Nat. Catal. 1, 12–22 (2018).
Noey, E. L. et al. Origins of stereoselectivity in evolved ketoreductases. Proc. Natl Acad. Sci. USA 112, E7065–E7072 (2015).
Xu, J. et al. Stereodivergent protein engineering of a lipase to access all possible stereoisomers of chiral esters with two stereocenters. J. Am. Chem. Soc. 141, 7934–7945 (2019).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Bayly, C. I., Cieplak, P., Cornell, W. D. & Kollman, P. A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97, 10269–10280 (1993).
Case, D. A. et al. Amber (2018); https://ambermd.org/
Kuhne, T. D. et al. CP2K: An electronic structure and molecular dynamics software package - Quickstep: efficient and accurate electronic structure calculations. J. Chem. Phys. 152, 194103 (2020).
VandeVondele, J. et al. QUICKSTEP: fast and accurate density functional calculations using a mixed Gaussian and plane waves approach. Comput. Phys. Commun. 167, 103–128 (2005).
Guidon, M., Hutter, J. & VandeVondele, J. Auxiliary density matrix methods for Hartree-Fock exchange calculations. J. Chem. Theory Comput. 6, 2348–2364 (2010).
Laio, A. & Parrinello, M. Escaping free-energy minima. Proc. Natl Acad. Sci. USA 99, 12562–12566 (2002).
Ensing, B., Laio, A., Parrinello, M. & Klein, M. L. A recipe for the computation of the free energy barrier and the lowest free energy path of concerted reactions. J. Phys. Chem. B 109, 6676–6687 (2005).
Frisch, M. J. et al. Gaussian 16 Rev. A.03 (2016); https://gaussian.com/
Marenich, A. V., Cramer, C. J. & Truhlar, D. G. Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J. Phys. Chem. B 113, 6378–6396 (2009).
Acknowledgements
This work was supported by the US Department of Energy (no. DE-SC0018420 to H.Z.), the National Natural Science Foundation of China (no. 2073077 to B.W.) and the National Key Research and Development Program of China (no. 2019YFA09005000 to J.Z.). NMR data were collected at the Carl R. Woese Institute for Genomic Biology Core, on a 600-MHz NMR funded by NIH grant no. S10-RR028833 (to H.Z.). We thank Codexis, Inc. for kindly sharing the amino acid sequence information of KRED-P2-D12, the staff of beamlines BL17U1 and BL19U1 of the Shanghai Synchrotron Radiation Facility and the National Center for Protein Science Shanghai for access and help with X-ray data collection.
Author information
Authors and Affiliations
Contributions
H.Z. coordinated the project. X.H. and H.Z. conceived the project and designed experiments. X.H. performed the majority of the experiments. J.F. and B.W. performed computational studies. J.C., X.Z. and J.Z. performed all structural biology studies. G.J. built the mutation library. W.H. contributed to scope investigation. X.H., B.W. and H.Z. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Catalysis thanks Chun-Jung Chen, Peng Tao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–47, Tables 1–16, discussions, HPLC spectra and NMR spectra.
Supplementary Data 1
Atomic coordinates of the optimized computational models.
Rights and permissions
About this article
Cite this article
Huang, X., Feng, J., Cui, J. et al. Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition. Nat Catal 5, 586–593 (2022). https://doi.org/10.1038/s41929-022-00777-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-022-00777-4
This article is cited by
-
Stereodivergent photobiocatalytic radical cyclization through the repurposing and directed evolution of fatty acid photodecarboxylases
Nature Chemistry (2024)
-
A light-driven enzymatic enantioselective radical acylation
Nature (2024)
-
Remote stereocontrol with azaarenes via enzymatic hydrogen atom transfer
Nature Chemistry (2024)
-
Direct visible-light-excited flavoproteins for redox-neutral asymmetric radical hydroarylation
Nature Catalysis (2023)
-
Photoenzymatic enantioselective intermolecular radical hydroamination
Nature Catalysis (2023)