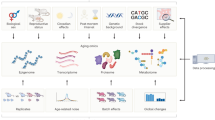
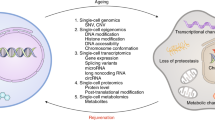
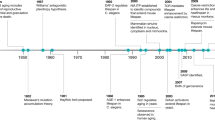
Abstract
The exponential scientific and technological progress during the past 30 years has favored the comprehensive characterization of aging processes with their multivariate nature, leading to the advent of Big Data in preclinical aging research. Spanning from molecular omics to organism-level deep phenotyping, Big Data demands large computational resources for storage and analysis, as well as new analytical tools and conceptual frameworks to gain novel insights leading to discovery. Systems biology has emerged as a paradigm that utilizes Big Data to gain insightful information enabling a better understanding of living organisms, visualized as multilayered networks of interacting molecules, cells, tissues and organs at different spatiotemporal scales. In this framework, where aging, health and disease represent emergent states from an evolving dynamic complex system, context given by, for example, strain, sex and feeding times, becomes paramount for defining the biological trajectory of an organism. Using bioinformatics and artificial intelligence, the systems biology approach is leading to remarkable advances in our understanding of the underlying mechanism of aging biology and assisting in creative experimental study designs in animal models. Future in-depth knowledge acquisition will depend on the ability to fully integrate information from different spatiotemporal scales in organisms, which will probably require the adoption of theories and methods from the field of complex systems. Here we review state-of-the-art approaches in preclinical research, with a focus on rodent models, that are leading to conceptual and/or technical advances in leveraging Big Data to understand basic aging biology and its full translational potential.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cohen, A. A. Complex systems dynamics in aging: new evidence, continuing questions. Biogerontology 17, 205–220 (2016).
Cohen, A. A. et al. A complex systems approach to aging biology. Nat. Aging 2, 580–591 (2022).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Zhavoronkov, A. et al. Artificial intelligence for aging and longevity research: recent advances and perspectives. Ageing Res. Rev. 49, 49–66 (2019).
Roser, M., Ritchie, H. & Mathieu, E. Technological change. Our World in Data https://ourworldindata.org/technological-change (2013).
Campisi, J. et al. From discoveries in ageing research to therapeutics for healthy ageing. Nature 571, 183–192 (2019).
Gauthier, J., Vincent, A. T., Charette, S. J. & Derome, N. A brief history of bioinformatics. Brief. Bioinform. 20, 1981–1996 (2019).
Barabasi, A.-L. Network Science (Cambridge Univ. Press, 2016).
Avchaciov, K. et al. Unsupervised learning of aging principles from longitudinal data. Nat. Commun. 13, 6529 (2022).
Xu, X., Hu, J., Lyu, X., Huang, H. & Cheng, X. Exploring the interdisciplinary nature of precision medicine: network analysis and visualization. JMIR Med. Inform. 9, e23562 (2021).
Sejnowski, T. J. The Deep Learning Revolution (The MIT Press, 2018).
Goh, W. W. B. & Wong, L. The birth of bio-data science: trends, expectations, and applications. Genomics Proteomics Bioinform. 18, 5–15 (2020).
Yang, J. H. et al. Loss of epigenetic information as a cause of mammalian aging. Cell 186, 305–326 e327 (2023).
Brunet, A., Goodell, M. A. & Rando, T. A. Ageing and rejuvenation of tissue stem cells and their niches. Nat. Rev. Mol. Cell Biol. 24, 45–62 (2023).
Acosta-Rodríguez, V. et al. Circadian alignment of early onset caloric restriction promotes longevity in male C57BL/6J mice. Science 376, 1192–1202 (2022).
Petr, M. A. et al. A cross-sectional study of functional and metabolic changes during aging through the lifespan in male mice. eLife 10, e62952 (2021).
Ristevski, B. & Chen, M. Big Data analytics in medicine and healthcare. J. Integr. Bioinform. 15, 20170030 (2018).
Meijering, E. A bird’s-eye view of deep learning in bioimage analysis. Comput. Struct. Biotechnol. J. 18, 2312–2325 (2020).
Voulodimos, A., Doulamis, N., Doulamis, A. & Protopapadakis, E. Deep learning for computer vision: a brief review. Comput. Intell. Neurosci. 2018, 7068349 (2018).
Suzuki, K. Overview of deep learning in medical imaging. Radiol. Phys. Technol. 10, 257–273 (2017).
Bermudez Contreras, E., Sutherland, R. J., Mohajerani, M. H. & Whishaw, I. Q. Challenges of a small world analysis for the continuous monitoring of behavior in mice. Neurosci. Biobehav. Rev. 136, 104621 (2022).
von Ziegler, L., Sturman, O. & Bohacek, J. Big behavior: challenges and opportunities in a new era of deep behavior profiling. Neuropsychopharmacology 46, 33–44 (2021).
Voikar, V. & Gaburro, S. Three pillars of automated home-cage phenotyping of mice: novel findings, refinement, and reproducibility based on literature and experience. Front. Behav. Neurosci. 14, 575434 (2020).
Perez-Riverol, Y. et al. Quantifying the impact of public omics data. Nat. Commun. 10, 3512 (2019).
Dato, S., Crocco, P., Rambaldi Migliore, N. & Lescai, F. Omics in a digital world: the role of bioinformatics in providing new insights into human aging. Front. Genet. 12, 689824 (2021).
Paigen, K. & Eppig, J. T. A mouse phenome project. Mamm. Genome 11, 715–717 (2000).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Haug, K. et al. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res 48, D440–D444 (2020).
Sud, M. et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–470 (2016).
Harris, P. A. et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biom. Inform. 42, 377–381 (2009).
Kahn, M. G., Eliason, B. B. & Bathurst, J. Quantifying clinical data quality using relative gold standards. AMIA Annu. Symp. Proc. 2010, 356–360 (2010).
Tomczak, K., Czerwińska, P. & Wiznerowicz, M. Review The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015, 68–77 (2015).
Clough, E. & Barrett, T. The Gene Expression Omnibus database. Methods Mol. Biol. 1418, 93–110 (2016).
Boussadi, A. & Zapletal, E. A. Fast Healthcare Interoperability Resources (FHIR) layer implemented over i2b2. BMC Med. Inform. Decis. Mak. 17, 120 (2017).
Xia, J., Psychogios, N., Young, N. & Wishart, D. S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660 (2009).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Zhao, K. & Rhee, S. Y. Interpreting omics data with pathway enrichment analysis. Trends. Genet. https://doi.org/10.1016/j.tig.2023.01.003 (2023).
Miller, R. A. et al. An aging Interventions Testing Program: study design and interim report. Aging Cell 6, 565–575 (2007).
Macchiarini, F., Miller, R. A., Strong, R., Rosenthal, N. & Harrison, D. E. in Handbook of the Biology of Aging (eds Musi, N. & Hornsby, P. J.) 219–235 (Academic Press, 2021).
Brown, S. D. M. & Moore, M. W. Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Dis. Model Mech. 5, 289–292 (2012).
Groza, T. et al. The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 51, D1038–D1045 (2023).
Churchill, G. A., Gatti, D. M., Munger, S. C. & Svenson, K. L. The Diversity Outbred mouse population. Mamm. Genome 23, 713–718 (2012).
Lee, P. J. et al. NIH SenNet Consortium to map senescent cells throughout the human lifespan to understand physiological health. Nat. Aging 2, 1090–1100 (2022).
Brown, S. D. M. et al. High-throughput mouse phenomics for characterizing mammalian gene function. Nat. Rev. Genet. 19, 357–370 (2018).
Palliyaguru, D. L. et al. Study of longitudinal aging in mice: presentation of experimental techniques. J. Gerontol. A 76, 552–560 (2021).
Successful Trajectories of Aging: Reserve and Resilience in RatS (STARRRS). National Institutes of Health https://www.nia.nih.gov/research/labs/about-irp/successful-trajectories-of-aging-reserve-and-resilience-in-rats#About (2022).
Evans, D. S. et al. Longitudinal functional study of murine aging: a resource for future study designs. JBMR Plus 5, e10466 (2021).
Kuo, P. L. et al. Longitudinal phenotypic aging metrics in the Baltimore Longitudinal Study of Aging. Nat. Aging 2, 635–643 (2022).
Smith, D. L. Jr. et al. Weight cycling increases longevity compared with sustained obesity in mice. Obesity 26, 1733–1739 (2018).
Ahadi, S. et al. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med. 26, 83–90 (2020).
Alfaras, I. et al. Empirical versus theoretical power and type I error (false-positive) rates estimated from real murine aging research data. Cell Rep. 36, 109560 (2021).
Kitano, H. Foundations of Systems Biology (The MIT Press, 2001).
Barabasi, A. L. & Albert, R. Emergence of scaling in random networks. Science 286, 509–512 (1999).
Lorusso, J. S., Sviderskiy, O. A. & Labunskyy, V. M. Emerging omics approaches in aging research. Antioxid. Redox Signal 29, 985–1002 (2018).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell 186, 243–278 (2023).
Wright, K. M. et al. Age and diet shape the genetic architecture of body weight in diversity outbred mice. eLife 11, e64329 (2022).
Dholaniya, P. S., Ghosh, S., Surampudi, B. R. & Kondapi, A. K. A knowledge driven supervised learning approach to identify gene network of differentially up-regulated genes during neuronal senescence in Rattus norvegicus. Biosystems 135, 9–14 (2015).
Wood, S. H., Craig, T., Li, Y., Merry, B. & de Magalhaes, J. P. Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age 35, 763–776 (2013).
de Magalhaes, J. P., Curado, J. & Church, G. M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881 (2009).
Li, N., Bates, D. J., An, J., Terry, D. A. & Wang, E. Up-regulation of key microRNAs, and inverse down-regulation of their predicted oxidative phosphorylation target genes, during aging in mouse brain. Neurobiol. Aging 32, 944–955 (2011).
Maes, O. C., An, J., Sarojini, H. & Wang, E. Murine microRNAs implicated in liver functions and aging process. Mech. Ageing Dev. 129, 534–541 (2008).
Khanna, A., Muthusamy, S., Liang, R., Sarojini, H. & Wang, E. Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging 3, 223–236 (2011).
Walther, D. M. & Mann, M. Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol. Cell Proteomics 10, M110 004523 (2011).
Houtkooper, R. H. et al. The metabolic footprint of aging in mice. Sci. Rep. 1, 134 (2011).
Tomas-Loba, A., Bernardes de Jesus, B., Mato, J. M. & Blasco, M. A. A metabolic signature predicts biological age in mice. Aging Cell 12, 93–101 (2013).
Rappley, I. et al. Lipidomic profiling in mouse brain reveals differences between ages and genders, with smaller changes associated with alpha-synuclein genotype. J. Neurochem. 111, 15–25 (2009).
Pak, H. H. et al. Fasting drives the metabolic, molecular and geroprotective effects of a calorie-restricted diet in mice. Nat. Metab. 3, 1327–1341 (2021).
Chondronasiou, D. et al. Multi-omic rejuvenation of naturally aged tissues by a single cycle of transient reprogramming. Aging Cell 21, e13578 (2022).
Aon, M. A. et al. Untangling determinants of enhanced health and lifespan through a multi-omics approach in mice. Cell Metab. 32, 100–116.e104 (2020).
Bernier, M. et al. Age-dependent impact of two exercise training regimens on genomic and metabolic remodeling in skeletal muscle and liver of male mice. NPJ Aging 8, 8 (2022).
Mitchell, S. J. et al. Daily fasting improves health and survival in male mice independent of diet composition and calories. Cell Metab. 29, 221–228.e223 (2019).
Benayoun, B. A. et al. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 29, 697–709 (2019).
Ori, A. et al. Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst. 1, 224–237 (2015).
Williams, E. G. et al. Multiomic profiling of the liver across diets and age in a diverse mouse population. Cell Syst. 13, 43–57.e46 (2022).
Sato, S. et al. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170, 664–677.e611 (2017).
Schaum, N. et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature 583, 596–602 (2020).
Shavlakadze, T. et al. Age-related gene expression signature in rats demonstrate early, late, and linear transcriptional changes from multiple tissues. Cell Rep. 28, 3263–3273.e3263 (2019).
Nie, C. et al. Distinct biological ages of organs and systems identified from a multi-omics study. Cell Rep. 38, 110459 (2022).
Xie, K. et al. Deep phenotyping and lifetime trajectories reveal limited effects of longevity regulators on the aging process in C57BL/6J mice. Nat. Commun. 13, 6830 (2022).
Ke, S. et al. Gut microbiota predicts healthy late-life aging in male mice. Nutrients 13, 3290 (2021).
Luo, D. et al. Gut microbiota combined with metabolomics reveals the metabolic profile of the normal aging process and the anti-aging effect of FuFang Zhenshu TiaoZhi(FTZ) in mice. Biomed. Pharmacother. 121, 109550 (2020).
Lee, B. P. et al. Changes in the expression of splicing factor transcripts and variations in alternative splicing are associated with lifespan in mice and humans. Aging Cell 15, 903–913 (2016).
Bou Sleiman, M. et al. Sex- and age-dependent genetics of longevity in a heterogeneous mouse population. Science 377, eabo3191 (2022).
Zhang, M. J., Pisco, A. O., Darmanis, S. & Zou, J. Mouse aging cell atlas analysis reveals global and cell type-specific aging signatures. eLife 10, e62293 (2021).
Palmer, D., Fabris, F., Doherty, A., Freitas, A. A. & de Magalhães, J. P. Ageing transcriptome meta-analysis reveals similarities and differences between key mammalian tissues. Aging 13, 3313–3341 (2021).
Ma, S. et al. Organization of the mammalian metabolome according to organ function, lineage specialization, and longevity. Cell Metab. 22, 332–343 (2015).
He, X., Memczak, S., Qu, J., Belmonte, J. C. I. & Liu, G.-H. Single-cell omics in ageing: a young and growing field. Nat. Metab. 2, 293–302 (2020).
Matteini, F., Mulaw, M. A. & Florian, M. C. Aging of the hematopoietic stem cell niche: new tools to answer an old question. Front. Immunol. 12, 738204 (2021).
Burlingame, E. A. et al. Toward reproducible, scalable, and robust data analysis across multiplex tissue imaging platforms. Cell Rep. Methods 1, 100053 (2021).
Amin, M. R., Yurovsky, A., Tian, Y. & Skiena, S. in Proc. 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics 254–259 (Association for Computing Machinery).
Kelley, D. R., Snoek, J. & Rinn, J. L. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 26, 990–999 (2016).
Asakura, T., Date, Y. & Kikuchi, J. Application of ensemble deep neural network to metabolomics studies. Anal. Chim. Acta 1037, 230–236 (2018).
Li, R., Li, L., Xu, Y. & Yang, J. Machine learning meets omics: applications and perspectives. Brief. Bioinform. 23, bbab460 (2022).
Marino, N. et al. Towards AI-driven longevity research: an overview. Front. Aging 4, 1057204 (2023).
Allen, W. E., Blosser, T. R., Sullivan, Z. A., Dulac, C. & Zhuang, X. Molecular and spatial signatures of mouse brain aging at single-cell resolution. Cell 186, 194–208.e118 (2023).
Bredikhin, D., Kats, I. & Stegle, O. MUON: multimodal omics analysis framework. Genome Biol. 23, 42 (2022).
Koehler Leman, J. et al. Ensuring scientific reproducibility in bio-macromolecular modeling via extensive, automated benchmarks. Nat. Commun. 12, 6947 (2021).
Wratten, L., Wilm, A. & Goke, J. Reproducible, scalable, and shareable analysis pipelines with bioinformatics workflow managers. Nat. Methods 18, 1161–1168 (2021).
Bellantuono, I. et al. A toolbox for the longitudinal assessment of healthspan in aging mice. Nat. Protoc. 15, 540–574 (2020).
Chen, Z. et al. Automated, high-dimensional evaluation of physiological aging and resilience in outbred mice. eLife 11, e72664 (2022).
Kempermann, G. et al. The individuality paradigm: automated longitudinal activity tracking of large cohorts of genetically identical mice in an enriched environment. Neurobiol. Dis. 175, 105916 (2022).
Acosta-Rodriguez, V. A., de Groot, M. H. M., Rijo-Ferreira, F., Green, C. B. & Takahashi, J. S. Mice under caloric restriction self-impose a temporal restriction of food intake as revealed by an automated feeder system. Cell Metab. 26, 267–277.e262 (2017).
Niemeyer, J. E. Telemetry for small animal physiology. Lab Anim. 45, 255–257 (2016).
Li, J. Y., Kuo, T. B. J. & Yang, C. C. H. Behaviour consistency is a sensitive tool for distinguishing the effects of aging on physical activity. Behav. Brain Res. 389, 112619 (2020).
Gill, S. & Panda, S. A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab. 22, 789–798 (2015).
Yang, C. C. & Hsu, Y. L. A review of accelerometry-based wearable motion detectors for physical activity monitoring. Sensors 10, 7772–7788 (2010).
Le Goallec, A. et al. Machine learning approaches to predict age from accelerometer records of physical activity at biobank scale. PLoS Digit. Health 2, e0000176 (2023).
Manero, A. et al. Improving disease prevention, diagnosis, and treatment using novel bionic technologies. Bioeng. Transl. Med. 8, e10359 (2023).
Alanazi, M. A. et al. Towards a low-cost solution for gait analysis using millimeter wave sensor and machine learning. Sensors 22, 5470 (2022).
Chakraborty, S., Aich, S., Joo, M. I., Sain, M. & Kim, H. C. A multichannel convolutional neural network architecture for the detection of the state of mind using physiological signals from wearable devices. J. Healthc. Eng. 2019, 5397814 (2019).
Zhang, W. B. & Pincus, Z. Predicting all-cause mortality from basic physiology in the Framingham Heart Study. Aging Cell 15, 39–48 (2016).
de Rezende, L. M. T. et al. Core temperature circadian rhythm across aging in spontaneously hypertensive rats. J. Therm. Biol. 97, 102807 (2021).
Axsom, J. E. et al. Acclimation to a thermoneutral environment abolishes age-associated alterations in heart rate and heart rate variability in conscious, unrestrained mice. GeroScience 42, 217–232 (2020).
Basso, A. et al. Circadian rhythms of body temperature and locomotor activity in aging BALB/c mice: early and late life span predictors. Biogerontology 17, 703–714 (2016).
Morrone, C. D., Tsang, A. A., Giorshev, S. M., Craig, E. E. & Yu, W. H. Concurrent behavioral and electrophysiological longitudinal recordings for in vivo assessment of aging. Front. Aging Neurosci. 14, 952101 (2022).
Raghunathan, S. et al. The design and hardware implementation of a low-power real-time seizure detection algorithm. J. Neural Eng. 6, 056005 (2009).
Martin-Montalvo, A. et al. Metformin improves healthspan and lifespan in mice. Nat. Commun. 4, 2192 (2013).
Martinez, P. et al. RAP1 protects from obesity through its extratelomeric role regulating gene expression. Cell Rep. 3, 2059–2074 (2013).
Arriola Apelo, S. I., Pumper, C. P., Baar, E. L., Cummings, N. E. & Lamming, D. W. Intermittent administration of rapamycin extends the life span of female C57BL/6J mice. J. Gerontol. A 71, 876–881 (2016).
Mercken, E. M. et al. SRT2104 extends survival of male mice on a standard diet and preserves bone and muscle mass. Aging Cell 13, 787–796 (2014).
Flesia, A. G., Nieto, P. S., Aon, M. A. & Kembro, J. M. in Computational Systems Biology in Medicine and Biotechnology: Methods and Protocols Vol. 2399 (eds Cortassa, S. & Aon, M. A.) 277–341 (Springer, 2022).
Baran, S. W. et al. Digital biomarkers enable automated, longitudinal monitoring in a mouse model of aging. J. Gerontol. A 76, 1206–1213 (2021).
Baran, S. W. et al. Emerging role of translational digital biomarkers within home cage monitoring technologies in preclinical drug discovery and development. Front. Behav. Neurosci. 15, 758274 (2021).
Bellman, R. E. Dynamic Programming (Dover Publications, 2003).
Di Germanio, C., Di Francesco, A., Bernier, M. & de Cabo, R. Yo-Yo dieting is better than none. Obesity 26, 1673–1673 (2018).
Mitchell, S. J. et al. Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 23, 1093–1112 (2016).
Nelson, P. G., Promislow, D. E. L. & Masel, J. Biomarkers for aging identified in cross-sectional studies tend to be non-causative. J. Gerontol. A 75, 466–472 (2020).
Prince, S. J. D. Computer Vision: Models, Learning, and Inference (Cambridge Univ. Press, 2012).
Dall’Ara, E. et al. Longitudinal imaging of the ageing mouse. Mech. Ageing Dev. 160, 93–116 (2016).
Balasubramani, V. et al. Roadmap on digital holography-based quantitative phase imaging. J. Imaging 7, 252 (2021).
Li, H. S. Genetic influences on susceptibility of the auditory system to aging and environmental factors. Scand. Audiol. Suppl. 36, 1–39 (1992).
Rosas, H. D., Feigin, A. S. & Hersch, S. M. Using advances in neuroimaging to detect, understand, and monitor disease progression in Huntington’s disease. NeuroRx 1, 263–272 (2004).
McConville, P., Moody, J. B. & Moffat, B. A. High-throughput magnetic resonance imaging in mice for phenotyping and therapeutic evaluation. Curr. Opin. Chem. Biol. 9, 413–420 (2005).
Minoshima, S. & Cross, D. In vivo imaging of axonal transport using MRI: aging and Alzheimer’s disease. Eur. J. Nucl. Med. Mol. Imaging 35, S89–92 (2008).
Mori, T. et al. Molecular imaging of dementia. Psychogeriatrics 12, 106–114 (2012).
Cao, L. et al. Positron emission tomography in animal models of tauopathies. Front. Aging Neurosci. 13, 761913 (2021).
Nagata, T. Macromolecular synthesis in the livers of aging mice as revealed by electron microscopic radioautography. Prog. Histochem. Cytochem. 45, 1–79 (2010).
Wassan, J. T., Zheng, H. & Wang, H. Role of deep learning in predicting aging-related diseases: a scoping review. Cells 10, 2924 (2021).
Liu, X., Song, L., Liu, S. & Zhang, Y. A review of deep-learning-based medical image segmentation methods. Sustainability 13, 1224 (2021).
Park, J. H., Seo, E., Choi, W. & Lee, S. J. Ultrasound deep learning for monitoring of flow-vessel dynamics in murine carotid artery. Ultrasonics 120, 106636 (2022).
Guimaraes, P. et al. Retinal aging in 3x Tg-AD mice model of Alzheimer’s disease. Front. Aging Neurosci. 14, 832195 (2022).
Haft-Javaherian, M. et al. Deep convolutional neural networks for segmenting 3D in vivo multiphoton images of vasculature in Alzheimer disease mouse models. PLoS ONE 14, e0213539 (2019).
Jo, T., Nho, K. & Saykin, A. J. Deep learning in Alzheimer’s disease: diagnostic classification and prognostic prediction using neuroimaging data. Front. Aging Neurosci. 11, 220 (2019).
Seo, S. Y. et al. Unified deep learning-based mouse brain MR segmentation: template-based individual brain positron emission tomography volumes-of-interest generation without spatial normalization in mouse Alzheimer model. Front. Aging Neurosci. 14, 807903 (2022).
Choi, S. et al. Automated characterisation of microglia in ageing mice using image processing and supervised machine learning algorithms. Sci. Rep. 12, 1806 (2022).
Tan, X., Su, A. T., Hajiabadi, H., Tran, M. & Nguyen, Q. Applying machine learning for integration of multi-modal genomics data and imaging data to quantify heterogeneity in tumour tissues. Methods Mol. Biol. 2190, 209–228 (2021).
Wang, Y. et al. Classification of mice hepatic granuloma microscopic images based on a deep convolutional neural network. Appl. Soft Comput. 74, 40–50 (2019).
Melgoza, I. P. et al. Development of a standardized histopathology scoring system using machine learning algorithms for intervertebral disc degeneration in the mouse model—an ORS spine section initiative. JOR Spine 4, e1164 (2021).
Kusumoto, D. et al. Automated deep learning-based system to identify endothelial cells derived from induced pluripotent stem cells. Stem Cell Rep. 10, 1687–1695 (2018).
Mathis, A. et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018).
Wade, L., Needham, L., McGuigan, P. & Bilzon, J. Applications and limitations of current markerless motion capture methods for clinical gait biomechanics. PeerJ 10, e12995 (2022).
Nath, T. et al. Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat Protoc 14, 2152–2176 (2019).
Hession, L. E., Sabnis, G. S., Churchill, G. A. & Kumar, V. A machine-vision-based frailty index for mice. Nat Aging 2, 756–766 (2022).
Schrack, J. A., Zipunnikov, V., Simonsick, E. M., Studenski, S. & Ferrucci, L. Rising energetic cost of walking predicts gait speed decline with aging. J. Gerontol. A 71, 947–953 (2016).
Bair, W.-N. et al. Of aging mice and men: gait speed decline is a translatable trait, with species-specific underlying properties. J. Gerontol. A 74, 1413–1416 (2019).
Cao, C. et al. Deep learning and its applications in biomedicine. Genomics Proteomics Bioinform. 16, 17–32 (2018).
Mathis, M. W. & Mathis, A. Deep learning tools for the measurement of animal behavior in neuroscience. Curr. Opin. Neurobiol. 60, 1–11 (2020).
van Dam, E. A., Noldus, L. P. J. J. & van Gerven, M. A. J. Deep learning improves automated rodent behavior recognition within a specific experimental setup. J. Neurosci. Methods 332, 108536 (2020).
Baker, G. T. & Sprott, R. L. Biomarkers of aging. Exp. Gerontol. 23, 223–239 (1988).
Brinkley, T. E. et al. Research priorities for measuring biologic age: summary and future directions from the Research Centers Collaborative Network Workshop. GeroScience 44, 2573–2583 (2022).
Wang, Q. et al. An evaluation of aging measures: from biomarkers to clocks. Biogerontology 24, 303–328 (2022).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, 3156 (2013).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013).
Galkin, F. et al. Biohorology and biomarkers of aging: current state-of-the-art, challenges and opportunities. Ageing Res. Rev. 60, 101050 (2020).
Rutledge, J., Oh, H. & Wyss-Coray, T. Measuring biological age using omics data. Nat. Rev. Genet. 23, 715–727 (2022).
Nielsen, J. L., Bakula, D. & Scheibye-Knudsen, M. Clinical trials targeting aging. Front. Aging 3, 820215 (2022).
Dubina, T. L., Dyundikova, V. A. & Zhuk, E. V. Biological age and its estimation. II. Assessment of biological age of albino rats by multiple regression analysis. Exp. Gerontol. 18, 5–18 (1983).
Dyundikova, V. A., Silvon, Z. K. & Dubina, T. L. Biological age and its estimation. I. Studies of some physiological parameters in albino rats and their validity as biological age tests. Exp. Gerontol. 16, 13–24 (1981).
Ludwig, F. C. & Smoke, M. E. The measurement of biological age. Exp. Aging Res. 6, 497–522 (1980).
Hofecker, G., Skalicky, M., Kment, A. & Niedermüller, H. Models of the biological age of the rat. I. A factor model of age parameters. Mech. Ageing Dev. 14, 345–359 (1980).
Skalicky, M., Hofecker, G., Kment, A. & Niedermüller, H. Models of the biological age of the rat. II. Multiple regression models in the study on influencing aging. Mech. Ageing Dev. 14, 361–377 (1980).
Levine, M. et al. A rat epigenetic clock recapitulates phenotypic aging and co-localizes with heterochromatin. eLife 9, e59201 (2020).
Schultz, M. B. et al. Age and life expectancy clocks based on machine learning analysis of mouse frailty. Nat. Commun. 11, 4618 (2020).
Bisset, E. S., Heinze-Milne, S., Grandy, S. A. & Howlett, S. E. Aerobic exercise attenuates frailty in aging male and female C57Bl/6 mice and effects systemic cytokines differentially by sex. J. Gerontol. A 77, 41–46 (2022).
Mach, J., Kane, A. E., Howlett, S. E., Sinclair, D. A. & Hilmer, S. N. Applying the AFRAID and FRIGHT clocks to novel preclinical mouse models of polypharmacy. J. Gerontol. A 77, 1304–1312 (2022).
Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 (2018).
Petkovich, D. A. et al. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metab. 25, 954–960.e956 (2017).
Meer, M. V., Podolskiy, D. I., Tyshkovskiy, A. & Gladyshev, V. N. A whole lifespan mouse multi-tissue DNA methylation clock. eLife 7, e40675 (2018).
Thompson, M. J. et al. A multi-tissue full lifespan epigenetic clock for mice. Aging 10, 2832–2854 (2018).
Coninx, E. et al. Hippocampal and cortical tissue-specific epigenetic clocks indicate an increased epigenetic age in a mouse model for Alzheimer’s disease. Aging 12, 20817–20834 (2020).
Trapp, A., Kerepesi, C. & Gladyshev, V. N. Profiling epigenetic age in single cells. Nat. Aging 1, 1189–1201 (2021).
Belsky, D. W. et al. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. eLife 9, e54870 (2020).
Freund, A. Untangling aging using dynamic, organism-level phenotypic networks. Cell Syst. 8, 172–181 (2019).
Mangul, S. et al. Systematic benchmarking of omics computational tools. Nat. Commun. 10, 1393 (2019).
Cortassa, S. & Aon, M. A. Computational Systems Biology in Medicine and Biotechnology: Methods and Protocols Vol. 2399 (Springer, 2022).
Han, Y. et al. Transcriptome features of striated muscle aging and predictability of protein level changes. Mol. Omics 17, 796–808 (2021).
Gligorijevic, V. & Przulj, N. Methods for biological data integration: perspectives and challenges. J. R. Soc. Interface 12, 20150571 (2015).
Rodosthenous, T., Shahrezaei, V. & Evangelou, M. Integrating multi-OMICS data through sparse canonical correlation analysis for the prediction of complex traits: a comparison study. Bioinformatics 36, 4616–4625 (2020).
Stahlschmidt, S. R., Ulfenborg, B. & Synnergren, J. Multimodal deep learning for biomedical data fusion: a review. Brief. Bioinform. 23, bbab569 (2022).
Jendoubi, T. Approaches to integrating metabolomics and multi-omics data: a primer. Metabolites 11, 184 (2021).
Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biol 18, 83 (2017).
Huang, S., Chaudhary, K. & Garmire, L. X. More is better: recent progress in multi-omics data integration methods. Front. Genet. 8, 84 (2017).
Chatsirisupachai, K., Lesluyes, T., Paraoan, L., Van Loo, P. & de Magalhaes, J. P. An integrative analysis of the age-associated multi-omic landscape across cancers. Nat. Commun. 12, 2345 (2021).
Aon, M. A., Bernier, M. & de Cabo, R. in Computational Systems Biology in Medicine and Biotechnology: Methods and Protocols Vol. 2399 (eds Cortassa, S. & Aon, M. A.) 193–218 (Springer, 2022).
Mattison, J. A. et al. Caloric restriction improves health and survival of rhesus monkeys. Nat. Commun. 8, 14063 (2017).
Mattison, J. A. et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature 489, 318–321 (2012).
Colman, R. J. et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325, 201–204 (2009).
Vijayakumar, S., Magazzu, G., Moon, P., Occhipinti, A. & Angione, C. in Computational Systems Biology in Medicine and Biotechnology: Methods and Protocols Vol. 2399 (eds Cortassa, S. & Aon, M. A.) 87–122 (Springer, 2022).
Bigan, E. et al. Genetic cooperativity in multi-layer networks implicates cell survival and senescence in the striatum of Huntington’s disease mice synchronous to symptoms. Bioinformatics 36, 186–196 (2020).
Ogris, C., Hu, Y., Arloth, J. & Muller, N. S. Versatile knowledge guided network inference method for prioritizing key regulatory factors in multi-omics data. Sci. Rep. 11, 6806 (2021).
Oh, S. et al. GenomicSuperSignature facilitates interpretation of RNA-seq experiments through robust, efficient comparison to public databases. Nat. Commun. 13, 3695 (2022).
Meyer, F. et al. Tutorial: assessing metagenomics software with the CAMI benchmarking toolkit. Nat. Protoc. 16, 1785–1801 (2021).
Das, S. & Mukhopadhyay, I. TiMEG: an integrative statistical method for partially missing multi-omics data. Sci. Rep. 11, 24077 (2021).
Tarazona, S. et al. Harmonization of quality metrics and power calculation in multi-omic studies. Nat. Commun. 11, 3092 (2020).
Eid, F. E. et al. Systematic auditing is essential to debiasing machine learning in biology. Commun. Biol. 4, 183 (2021).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Succi, S. & Coveney, P. V. Big data: the end of the scientific method? Phil. Trans. R. Soc. A 377, 20180145 (2019).
Parkinson, H. et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 35, D747–D750 (2007).
GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Vizcaino, J. A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014).
Deutsch, E. W. et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 51, D1539–D1548 (2023).
Perez-Riverol, Y. et al. Discovering and linking public omics data sets using the Omics Discovery Index. Nat. Biotechnol. 35, 406–409 (2017).
Schaum, N. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
Tabula Muris Consortium A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020).
Jarmusch, A. K. et al. ReDU: a framework to find and reanalyze public mass spectrometry data. Nat. Methods 17, 901–904 (2020).
Aging Atlas Consortium Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res. 49, D825–D830 (2020).
Samad, M., Agostinelli, F., Sato, T., Shimaji, K. & Baldi, P. CircadiOmics: circadian omic web portal. Nucleic Acids Res. 50, W183–W190 (2022).
Ma, L. et al. Database Commons: a catalog of worldwide biological databases. Genomics Proteomics Bioinform. https://doi.org/10.1016/j.gpb.2022.12.004 (2022).
Pilarczyk, M. et al. Connecting omics signatures and revealing biological mechanisms with iLINCS. Nat. Commun. 13, 4678 (2022).
Gao, Y. et al. AgingBank: a manually curated knowledgebase and high-throughput analysis platform that provides experimentally supported multi-omics data relevant to aging in multiple species. Brief. Bioinform. 23, bbac438 (2022).
Richardson, J. E., Eppig, J. T. & Nadeau, J. H. Building an integrated mouse genome database: a view from the front line. IEEE Eng. Med. Biol. Mag. 14, 718–724 (1995).
Ringwald, M. et al. Mouse Genome Informatics (MGI): latest news from MGD and GXD. Mamm. Genome 33, 4–18 (2022).
Twigger, S. et al. Rat Genome Database (RGD): mapping disease onto the genome. Nucleic Acids Res. 30, 125–128 (2002).
Smith, J. R. et al. The Year of the Rat: The Rat Genome Database at 20: a multi-species knowledgebase and analysis platform. Nucleic Acids Res. 48, D731–D742 (2020).
de Magalhães, J. P., Costa, J. & Toussaint, O. HAGR: the human ageing genomic resources. Nucleic Acids Res. 33, D537–D543 (2005).
Luo, Y. et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 48, D882–D889 (2020).
Consortium, T. E. P. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640 (2004).
Craig, T. et al. The Digital Ageing Atlas: integrating the diversity of age-related changes into a unified resource. Nucleic Acids Res. 43, D873–D878 (2015).
Peterson, K. A. & Murray, S. A. Progress towards completing the mutant mouse null resource. Mamm. Genome 33, 123–134 (2022).
Gallagher, M. et al. What are the threats to successful brain and cognitive aging? Neurobiol. Aging 83, 130–134 (2019).
McQuail, J. A. et al. Cognitive reserve in model systems for mechanistic discovery: the importance of longitudinal studies. Front. Aging Neurosci. 12, 607685 (2021).
Pang, Z. et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 49, W388–W396 (2021).
Mina, A. I. et al. CalR: a web-based analysis tool for indirect calorimetry experiments. Cell Metab. 28, 656–666.e651 (2018).
Lauer, J. et al. Multi-animal pose estimation, identification and tracking with DeepLabCut. Nat. Methods 19, 496–504 (2022).
Zierer, J., Menni, C., Kastenmuller, G. & Spector, T. D. Integration of ‘omics’ data in aging research: from biomarkers to systems biology. Aging Cell 14, 933–944 (2015).
Deng, G. et al. Targeting cathepsin B by cycloastragenol enhances antitumor immunity of CD8 T cells via inhibiting MHC-I degradation. J. Immunother. Cancer 10, e004874 (2022).
Lu, R. J. et al. Multi-omic profiling of primary mouse neutrophils predicts a pattern of sex and age-related functional regulation. Nat. Aging 1, 715–733 (2021).
Wang, Y., Liu, L., Song, Y., Yu, X. & Deng, H. Unveiling E2F4, TEAD1 and AP-1 as regulatory transcription factors of the replicative senescence program by multi-omics analysis. Protein Cell 13, 742–759 (2022).
Mahmoudi, S. et al. Heterogeneity in old fibroblasts is linked to variability in reprogramming and wound healing. Nature 574, 553–558 (2019).
Benjamin, D. I. et al. Multiomics reveals glutathione metabolism as a driver of bimodality during stem cell aging. Cell Metab. 35, 472–486.e476 (2023).
Xie, S. et al. Aging and obesity prime the methylome and transcriptome of adipose stem cells for disease and dysfunction. FASEB J. 37, e22785 (2023).
Nodari, A. et al. Interferon regulatory factor 7 impairs cellular metabolism in aging adipose-derived stromal cells. J. Cell Sci. 134, jcs256230 (2021).
Moudra, A. et al. Phenotypic and clonal stability of antigen-inexperienced memory-like T cells across the genetic background, hygienic status, and aging. J. Immunol. 206, 2109–2121 (2021).
Baptista, L. C. et al. Multiomics profiling of the impact of an angiotensin (1–7)-expressing probiotic combined with exercise training in aged male rats. J. Appl. Physiol. 134, 1135–1153 (2023).
Yang, G. et al. The essential roles of FXR in diet and age influenced metabolic changes and liver disease development: a multi-omics study. Biomark. Res. 11, 20 (2023).
Roberts, B. M. et al. Effects of an exogenous ketone ester using multi-omics in skeletal muscle of aging C57BL/6J male mice. Front. Nutr. 9, 1041026 (2022).
Lu, Y. et al. Multi-omics analysis reveals neuroinflammation, activated glial signaling, and dysregulated synaptic signaling and metabolism in the hippocampus of aged mice. Front. Aging Neurosci. 14, 964429 (2022).
Jiang, X. et al. An integrated multi-omics approach revealed the regulation of melatonin on age-dependent mitochondrial function impair and lipid dyshomeostasis in mice hippocampus. Pharmacol. Res. 179, 106210 (2022).
Roichman, A. et al. Restoration of energy homeostasis by SIRT6 extends healthy lifespan. Nat. Commun. 12, 3208 (2021).
Currais, A. et al. A comprehensive multiomics approach toward understanding the relationship between aging and dementia. Aging 7, 937–955 (2015).
Zhang, D. et al. Spatial epigenome-transcriptome co-profiling of mammalian tissues. Nature 616, 113–122 (2023).
Hernando-Herraez, I. et al. Ageing affects DNA methylation drift and transcriptional cell-to-cell variability in mouse muscle stem cells. Nat. Commun. 10, 4361 (2019).
Al-Amrani, S., Al-Jabri, Z., Al-Zaabi, A., Alshekaili, J. & Al-Khabori, M. Proteomics: concepts and applications in human medicine. World J. Biol. Chem. 12, 57–69 (2021).
Haas, R. et al. Designing and interpreting ‘multi-omic’ experiments that may change our understanding of biology. Curr. Opin. Syst. Biol. 6, 37–45 (2017).
Wei, P. et al. Urinary metabolomic and proteomic analyses in a mouse model of prostatic inflammation. Urine 1, 17–23 (2019).
Feng, Q. et al. The anti-aging effects of Renshen Guben on thyrotoxicosis mice: improving immunosenescence, hypoproteinemia, lipotoxicity, and intestinal flora. Front. Immunol. 13, 983501 (2022).
Labunskyy, V. M. & Gladyshev, V. N. Role of reactive oxygen species-mediated signaling in aging. Antioxid. Redox Signal 19, 1362–1372 (2013).
Miyajima, M. et al. Leucine-rich α2-glycoprotein is a novel biomarker of neurodegenerative disease in human cerebrospinal fluid and causes neurodegeneration in mouse cerebral cortex. PLoS ONE 8, e74453 (2013).
Porpiglia, E. et al. Elevated CD47 is a hallmark of dysfunctional aged muscle stem cells that can be targeted to augment regeneration. Cell Stem Cell 29, 1653–1668.e1658 (2022).
Vafadarnejad, E. et al. Dynamics of cardiac neutrophil diversity in murine myocardial infarction. Circ. Res. 127, e232–e249 (2020).
Han, X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 12, 668–679 (2016).
Bauer, K. C. et al. Dietary intervention reverses fatty liver and altered gut microbiota during early-life undernutrition. mSystems 5, e00499-20 (2020).
Zhang, B. et al. Gut microbiota dysbiosis induced by decreasing endogenous melatonin mediates the pathogenesis of Alzheimer’s disease and obesity. Front. Immunol. 13, 900132 (2022).
Kabra, M., Robie, A. A., Rivera-Alba, M., Branson, S. & Branson, K. JAABA: interactive machine learning for automatic annotation of animal behavior. Nat. Methods 10, 64–67 (2013).
Machado, A. S., Darmohray, D. M., Fayad, J., Marques, H. G. & Carey, M. R. A quantitative framework for whole-body coordination reveals specific deficits in freely walking ataxic mice. eLife 4, e07892 (2015).
Graving, J. M. et al. DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning. eLife 8, e47994 (2019).
Pereira, T. D. et al. Fast animal pose estimation using deep neural networks. Nat. Methods 16, 117–125 (2019).
Romero-Ferrero, F., Bergomi, M. G., Hinz, R. C., Heras, F. J. H. & de Polavieja, G. G. idtracker.ai: tracking all individuals in small or large collectives of unmarked animals. Nat. Methods 16, 179–182 (2019).
Arac, A., Zhao, P., Dobkin, B. H., Carmichael, S. T. & Golshani, P. DeepBehavior: a deep learning toolbox for automated analysis of animal and human behavior imaging data. Front. Syst. Neurosci. 13, 20 (2019).
Wiltschko, A. B. et al. Revealing the structure of pharmacobehavioral space through motion sequencing. Nat. Neurosci. 23, 1433–1443 (2020).
Karashchuk, P. et al. Anipose: a toolkit for robust markerless 3D pose estimation. Cell Rep. 36, 109730 (2021).
Pereira, T. D. et al. SLEAP: a deep learning system for multi-animal pose tracking. Nat. Methods 19, 486–495 (2022).
Wang, T. et al. Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 18, 57 (2017).
Stubbs, T. M. et al. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 18, 68 (2017).
Antoch, M. P. et al. Physiological frailty index (PFI): quantitative in-life estimate of individual biological age in mice. Aging 9, 615–626 (2017).
Han, Y. et al. Epigenetic age-predictor for mice based on three CpG sites. eLife 7, e37462 (2018).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Iijima, H. et al. Meta-analysis integrated with multi-omics data analysis to elucidate pathogenic mechanisms of age-related knee osteoarthritis in mice. J. Gerontol. A 77, 1321–1334 (2022).
Snel, B., Lehmann, G., Bork, P. & Huynen, M. A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28, 3442–3444 (2000).
Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023).
D’Amico, D. et al. The RNA-binding protein PUM2 impairs mitochondrial dynamics and mitophagy during aging. Mol. Cell 73, 775–787.e710 (2019).
Dennis, G. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, R60 (2003).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Otasek, D., Morris, J. H., Boucas, J., Pico, A. R. & Demchak, B. Cytoscape Automation: empowering workflow-based network analysis. Genome Biol. 20, 185 (2019).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9, 559 (2008).
Cottret, L. et al. MetExplore: a web server to link metabolomic experiments and genome-scale metabolic networks. Nucleic Acids Res. 38, W132–137 (2010).
Rinschen, M. M. et al. Metabolic rewiring of the hypertensive kidney. Sci. Signal 12, eaax9760 (2019).
Liu, T. et al. PaintOmics 4: new tools for the integrative analysis of multi-omics datasets supported by multiple pathway databases. Nucleic Acids Res. 50, W551–W559 (2022).
Kamburov, A., Cavill, R., Ebbels, T. M., Herwig, R. & Keun, H. C. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics 27, 2917–2918 (2011).
Kramer, A., Green, J., Pollard, J. Jr. & Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014).
Zhou, G. et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 47, W234–W241 (2019).
Arneson, D., Bhattacharya, A., Shu, L., Mäkinen, V.-P. & Yang, X. Mergeomics: a web server for identifying pathological pathways, networks, and key regulators via multidimensional data integration. BMC Genomics 17, 722 (2016).
Ding, J. et al. Mergeomics 2.0: a web server for multi-omics data integration to elucidate disease networks and predict therapeutics. Nucleic Acids Res. 49, W375–W387 (2021).
Gao, S., Casey, A. E., Sargeant, T. J. & Mäkinen, V.-P. Genetic variation within endolysosomal system is associated with late-onset Alzheimer’s disease. Brain 141, 2711–2720 (2018).
Rohart, F., Gautier, B., Singh, A. & Le Cao, K. A. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752 (2017).
Guttà, C., Morhard, C. & Rehm, M. Applying a GAN-based classifier to improve transcriptome-based prognostication in breast cancer. PLoS Comput. Biol. 19, e1011035 (2023).
Hinshaw, S. J., Lee, A. H. Y., Gill, E. E. & Hancock, R. E. W. MetaBridge: enabling network-based integrative analysis via direct protein interactors of metabolites. Bioinformatics 34, 3225–3227 (2018).
Bell, H. N. et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell 40, 185–200 e186 (2022).
Uppal, K., Ma, C., Go, Y. M., Jones, D. P. & Wren, J. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics 34, 701–702 (2018).
Koh, H. W. L. et al. iOmicsPASS: network-based integration of multiomics data for predictive subnetwork discovery. NPJ Syst. Biol. Appl. 5, 22 (2019).
Baum, A. & Vermue, L. Multiblock PLS: block dependent prediction modeling for Python. J. Open Source Softw. 4, 1190 (2019).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Mallick, H. et al. Predictive metabolomic profiling of microbial communities using amplicon or metagenomic sequences. Nat. Commun. 10, 3136 (2019).
Jun, S. R. et al. Multi-omic analysis reveals different effects of sulforaphane on the microbiome and metabolome in old compared to young mice. Microorganisms 8, 1500 (2020).
McIntyre, L. M. et al. GAIT-GM: Galaxy tools for modeling metabolite changes as a function of gene expression. Preprint at bioRxiv https://doi.org/10.1101/2020.12.25.424407 (2020).
McIntyre, L. M. et al. GAIT-GM integrative cross-omics analyses reveal cholinergic defects in a C. elegans model of Parkinson’s disease. Sci Rep 12, 3268 (2022).
Canzler, S. & Hackermuller, J. multiGSEA: a GSEA-based pathway enrichment analysis for multi-omics data. BMC Bioinform. 21, 561 (2020).
Song, X. et al. Multi-omics characterization of type 2 diabetes mellitus-induced cognitive impairment in the db/db mouse model. Molecules 27, 1904 (2022).
Tal, O., Selvaraj, G., Medina, S., Ofaim, S. & Freilich, S. NetMet: a network-based tool for predicting metabolic capacities of microbial species and their interactions. Microorganisms 8, 840 (2020).
Zhou, G., Ewald, J. & Xia, J. OmicsAnalyst: a comprehensive web-based platform for visual analytics of multi-omics data. Nucleic Acids Res. 49, W476–W482 (2021).
Makarious, M. B. et al. GenoML: automated machine learning for genomics. Preprint at https://doi.org/10.48550/arXiv.2103.03221 (2021).
Mechteridis, K., Lauber, M., Baumbach, J. & List, M. KeyPathwayMineR: de novo pathway enrichment in the R ecosystem. Front. Genet. 12, 812853 (2021).
Ghanat Bari, M., Ung, C. Y., Zhang, C., Zhu, S. & Li, H. Machine learning-assisted network inference approach to identify a new class of genes that coordinate the functionality of cancer networks. Sci. Rep. 7, 6993 (2017).
Bodein, A., Scott-Boyer, M. P., Perin, O., Le Cao, K. A. & Droit, A. Interpretation of network-based integration from multi-omics longitudinal data. Nucleic Acids Res. 50, e27 (2022).
Noecker, C., Eng, A., Muller, E. & Borenstein, E. MIMOSA2: a metabolic network-based tool for inferring mechanism-supported relationships in microbiome-metabolome data. Bioinformatics 38, 1615–1623 (2022).
Pun, F. W. et al. Hallmarks of aging-based dual-purpose disease and age-associated targets predicted using PandaOmics AI-powered discovery engine. Aging 14, 2475–2506 (2022).
Harbig, T. A., Fratte, J., Krone, M. & Nieselt, K. OmicsTIDE: interactive exploration of trends in multi-omics data. Bioinform. Adv. 3, vbac093 (2023).
Hopfield, J. J. Neural networks and physical systems with emergent collective computational abilities. Proc. Natl Acad. Sci. USA 79, 2554–2558 (1982).
Rosenblatt, F. Principles of Neurodynamics: Perceptrons and the Theory of Brain Mechanisms (Spartan, 1962).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. in Advances in Neural Information Processing Systems 25 (eds Pereira, F., Burges, C. J., Bottou, L. & Weinberger, K. Q.) 1097–1105 (Advances in Neural Information Processing Systems, 2012)
Erdos, P. & Renyi, A. On random graphs I. Publ. Math. Debrecen 6, 290–297 (1959).
Granovetter, M. S. The strenght of weak ties. Am. J. Sociol. 78, 1360–1380 (1973).
Watts, D. J. & Strogatz, S. H. Collective dynamics of ‘small-world’ networks. Nature 393, 440–442 (1998).
Mitchell, M. Artificial Intelligence: A Guide for Thinking Humans (Farrar, Strauss and Giroux, 2019).
Noble, R., Tasaki, K., Noble, P. J. & Noble, D. Biological relativity requires circular causality but not symmetry of causation: so, where, what and when are the boundaries? Front. Physiol. 10, 827–827 (2019).
Fernandez, M. E. et al. Context- and scale-dependent effects of thymol bioactivity on biological networks: contributions from quail under heat stress. Preprint at bioRxiv https://doi.org/10.1101/2022.06.10.495659 (2022).
Cortassa, S., Aon, M. A., Iglesias, A. A., Aon, J. C. & Lloyd, D. An Introduction to Metabolic and Cellular Engineering 2nd edn (World Scientific Publishing, 2012).
Vidal, M. A unifying view of 21st century systems biology. FEBS Lett. 583, 3891–3894 (2009).
Von Bertalanffy, L. The theory of open systems in physics and biology. Science 111, 23–29 (1950).
Aon, M. A., Lloyd, D. & Saks, V. in Systems Biology of Metabolic and Signaling Networks. Energy, Mass and Information Transfer Vol. 16 (eds Aon, M. A., Saks, V. & Schlattner, U.) 3–17 (Springer, 2014).
Noble, D. & Boyd, C. A. R. in Logic of Life: The Challenge of Integrative Physiology (eds Boyd, C. A. R. & Noble, D.) 1–13 (Oxford Univ. Press, 1993).
MacEachern, S. J. & Forkert, N. D. Machine learning for precision medicine. Genome 64, 416–425 (2020).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Big data and analytics support. IBM https://www.ibm.com/docs/en/spectrum-scale-bda?topic=big-data-analytics-support (2023).
Wilder-James, E. What is big data? An introduction to the big data landscape. O’Reilly https://www.oreilly.com/radar/what-is-big-data/ (2012).
Acknowledgements
M.A.A. is an employee of Kelly Government Solutions. This work was supported by the Intramural Research Program of the National Institute on Aging/NIH.
Author information
Authors and Affiliations
Contributions
All authors were responsible for conceptualization and methodology. M.E.F., J.M.-R. and M.A.A. were responsible for data collection and writing the original draft. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Lab Animal thanks Dan Ehninger, Simon Melov and João Pedro de Magalhães for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Sheet 1: Supplementary Table 1. Additional multi-omics studies in preclinical aging research in rodent models. Sheet 2: Supplementary Table 2. Number of PubMed articles retrieved with keywords representing the major driving forces in Fig. 1a, at global or preclinical aging research level.
Rights and permissions
About this article
Cite this article
Fernandez, M.E., Martinez-Romero, J., Aon, M.A. et al. How is Big Data reshaping preclinical aging research?. Lab Anim 52, 289–314 (2023). https://doi.org/10.1038/s41684-023-01286-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41684-023-01286-y