Abstract
Adoptive cell transfer between genetically identical hosts relies on the use of a congenic marker to distinguish the donor cells from the host cells. CD45, a glycoprotein expressed by all hematopoietic cells, is one of the main congenic markers used because its two isoforms, CD45.1 and CD45.2, can be discriminated by flow cytometry. As a consequence, C57BL/6J (B6; CD45.2) and B6.SJL-Ptprca Pepcb/BoyJ (B6.SJL; CD45.1) mice are widely used in adoptive cell transfer experiments, under the presumption that they differ only at the CD45 (Ptprc) locus. However, recent studies have identified genetic variations between these congenic strains and have notably highlighted a differential expression of cathepsin E (CTSE). The B6.SJL mouse presents a number of functional differences in hematopoietic stem cell engraftment potential and immune cell numbers compared with the B6 mouse. In this study, we showed that B6 and B6.SJL mice also differ in their CD8+ T cell compartment and CD8+ T cell responses to viral infection. We identified Ctse as the most differentially expressed gene between CD8+ T cells of B6 and B6.SJL and demonstrated that the differences reported between these two mouse strains are not due to CTSE. Finally, using CRISPR–Cas9 genome editing, we generated a CD45.1-expressing B6 mouse by inserting one nucleotide mutation (A904G) leading to an amino acid change (K302E) in the Ptprc gene of the B6 mouse. We showed that this new B6-Ptprcem(K302E)Jmar/J mouse resolves the experimental biases reported between the B6 and B6.SJL mouse lines and should thus represent the new gold standard for adoptive cell transfer experiments in B6.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The mouse models will be available through EMMA (https://www.infrafrontier.eu/emma/) once the paper is published. The microarray data have been deposited to the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE86601. Source data are provided with this paper.
References
Shen, F. W. et al. Cloning of Ly-5 cDNA. Proc. Natl Acad. Sci. USA 82, 7360–7363 (1985).
Waterstrat, A., Liang, Y., Swiderski, C. F., Shelton, B. J. & Van Zant, G. Congenic interval of CD45/Ly-5 congenic mice contains multiple genes that may influence hematopoietic stem cell engraftment. Blood 115, 408–417 (2010).
Chisolm, D. A. et al. Defining genetic variation in widely used congenic and backcrossed mouse models reveals varied regulation of genes important for immune responses. Immunity 51, 155–168.e5 (2019).
Basu, S., Ray, A. & Dittel, B. N. Differential representation of B cell subsets in mixed bone marrow chimera mice due to expression of allelic variants of CD45 (CD45.1/CD45.2). J. Immunol. Methods 396, 163–167 (2013).
Mercier, F. E., Sykes, D. B. & Scadden, D. T. Single targeted exon mutation creates a true congenic mouse for competitive hematopoietic stem cell transplantation: the C57BL/6-CD45.1 STEM mouse. Stem Cell Rep. 6, 985–992 (2016).
Jafri, S., Moore, S. D., Morrell, N. W. & Ormiston, M. L. A sex-specific reconstitution bias in the competitive CD45.1/CD45.2 congenic bone marrow transplant model. Sci. Rep. 7, 3495 (2017).
Jang, Y. et al. Cutting edge: check your mice—a point mutation in the Ncr1 locus identified in CD45.1 congenic mice with consequences in mouse susceptibility to infection. J. Immunol. 200, 1982–1987 (2018).
Gray, E. E. et al. Deficiency in IL-17-committed Vγ4+ γδ T cells in a spontaneous Sox13-mutant CD45.1+ congenic mouse substrain provides protection from dermatitis. Nat. Immunol. 14, 584–592 (2013).
Tulone, C., Tsang, J., Prokopowicz, Z., Grosvenor, N. & Chain, B. Natural cathepsin E deficiency in the immune system of C57BL/6J mice. Immunogenetics 59, 927–935 (2007).
Grau, M. et al. Antigen-induced but not innate memory CD8 T cells express NKG2D and are recruited to the lung parenchyma upon viral infection. J. Immunol. 200, 3635–3646 (2018).
Zebedee, S. L., Barritt, D. S. & Raschke, W. C. Comparison of mouse Ly5a. and Ly5b leucocyte common antigen alleles. Dev. Immunol. 12, 243–254 (1991).
Kornete, M., Marone, R. & Jeker, L. T. Highly efficient and versatile plasmid-based gene editing in primary T cells. J. Immunol. 200, 2489–2501 (2018).
Concordet, J.-P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Chain, B. M. et al. The expression and function of cathepsin E in dendritic cells. J. Immunol. 174, 1791–1800 (2005).
Kakehashi, H. et al. Differential regulation of the nature and functions of dendritic cells and macrophages by cathepsin E. J. Immunol. 179, 5728–5737 (2007).
Pilzner, C. et al. Allergic airway inflammation in mice deficient for the antigen-processing protease cathepsin E. Int. Arch. Allergy Immunol. 159, 367–383 (2012).
Teixeira, M. et al. Electroporation of mice zygotes with dual guide RNA/Cas9 complexes for simple and efficient cloning-free genome editing. Sci. Rep. 8, 474 (2018).
Simon, M. M. et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 14, R82 (2013).
Hiramatsu, S. Regulation of Cathepsin E gene expression by the transcription factor Kaiso in MRL/lpr mice derived CD4+ T cells. Sci. Rep. 9, 3054 (2019).
Tsukuba, T. Association of cathepsin E deficiency with development of atopic dermatitis. J. Biochem. 134, 893–902 (2003).
Mengwasser, J. et al. Cathepsin E deficiency ameliorates graft-versus-host disease and modifies dendritic cell motility. Front. Immunol. 8, 203 (2017).
Jubin, V. et al. T inflammatory memory CD8 T cells participate to antiviral response and generate secondary memory cells with an advantage in XCL1 production. Immunol. Res. 52, 284–293 (2012).
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989 (2015).
Richardson, C. D., Ray, G. J., DeWitt, M. A., Curie, G. L. & Corn, J. E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR–Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339–344 (2016).
Renaud, J.-B. et al. Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR–Cas9 nucleases. Cell Rep. 14, 2263–2272 (2016).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Brinza, L. et al. Immune signatures of protective spleen memory CD8 T cells. Sci. Rep. 6, 37651 (2016).
Acknowledgements
We acknowledge the contribution of AniRA-Cytometrie and AniRA-PBES, of the SFR BioSciences (UAR3444/CNRS, US8/Inserm, Ecole Normale Supérieure de Lyon, Université de Lyon). We acknowledge the contributions of the CELPHEDIA Infrastructure (http://www.celphedia.eu/). The project was funded by the intramural CIRI grant: AO-4-2018.
Author information
Authors and Affiliations
Contributions
D.L., J.M., S.D. and S.M. designed the study. D.L., M.D. and S.D. performed the experiment. C.A., F.H. and M.T. produced the mouse models. D.L., J.M. and S.M. wrote the manuscript. B.C. and Y.L. provided helpful discussions. S.D., Y.L. and B.C. reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Lab Animal thanks Bonnie Dittel and Maegan Capitano for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Ctse mRNA expression levels in various immune cell types from Immgen database.
The Immunological Genome Project (ImmGen) provides an atlas of mRNA signatures across the immune system. The figure represents the Ctse mRNA levels from various immune cell types isolated from a mouse with a B6 or a B6.SJL background.
Extended Data Fig. 2 Analysis of potential off-target sites for mutation A insertion.
a, The potential off-target (OT) sites for mutA-guide RNA were determined using CRISPOR software. Weaker NAG or NGA PAM are also taken into account even if corresponding OT are less likely to happen. Potential OT loci were PCR-amplified and sequenced using genomic DNA from founder B6.mutA F1 mouse and compared to B6 control. b, Corresponding PCR gels are shown. O’GeneRuler 100 bp Plus DNA Ladder was used as a ladder (L). c, Sequence alignments of potential OT regions for B6.mutA and B6 mice. Sequence targeted by the guide RNA is in blue, PAM sequences are in orange (NGG, NGA or NAG) and nucleotide differences between B6 and B6.mutA are in red.
Extended Data Fig. 3 Sequencing trace data of potential off-target sites.
Chromatograms of sequenced PCR products for each potential mutA OT sites and associated double stranded sequences are represented for B6.mutA and B6 mice. Sequence targeted by the guide RNA is highlighted in blue and PAM sequence in orange. Nucleotide differences are highlighted with a red square.
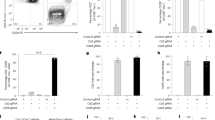
Extended Data Fig. 4 B6 and B6.mutA mice have similar bone marrow reconstitution potential.
Irradiated B6/B6.SJL heterozygous mice received a 1:1 mixture of B6.mutA or B6.SJL and B6 bone marrow cells. The proportions of B and T cells within CD45.1 and CD45.2 donor cells was determined 17 weeks after transfer in the spleen by flow cytometry. Results are expressed as the median ± range (n = 5 mice/group, one of two representative experiments). The statistical significance of differences was determined by Mann-Whitney test (*P < 0.05, **P < 0,01).
Extended Data Fig. 5 Generation of B6.SJL.CTSE-KO mice.
B6.SJL.CTSE-KO mouse was generated by CRIPSR editing. A 83 bp deletion was made in exon 3 of the Ctse gene in B6.SJL mouse using two guide RNAs simultaneously. a, The targeted Ctse gene sequence is represented with guide RNA sequences highlighted in red, PAM sequences in blue and primer sequences in grey. b, DNA Sanger sequencing of the targeted Ctse gene region for both B6.SJL.CTSE-KO and B6.SJL.CTSE-WT mice (sequences and trace data, antisense strand).
Supplementary information
Supplementary Table 1
List of oligonucleotide sequences.
Source data
Source Data Extended Data Fig. 2
Unprocessed gels related to Extended Data Fig. 2.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Laubreton, D., Djebali, S., Angleraux, C. et al. Generation of a C57BL/6J mouse strain expressing the CD45.1 epitope to improve hematopoietic stem cell engraftment and adoptive cell transfer experiments. Lab Anim 52, 324–331 (2023). https://doi.org/10.1038/s41684-023-01275-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41684-023-01275-1