Abstract
Hypothesizing after the results are known (HARK) has been disparaged as data dredging, and safeguards including hypothesis preregistration and statistically rigorous oversight have been recommended. Despite potential drawbacks, HARK has deepened thinking about complex causal processes. Some of the HARK precautions can conflict with the modern reality of researchers’ obligations to use big, ‘organic’ data sources—from high-throughput genomics to social media streams. We here propose a HARK-solid, reproducible inference framework suitable for big data, based on models that represent formalization of hypotheses. Reproducibility is attained by employing two levels of model validation: internal (relative to data collated around hypotheses) and external (independent to the hypotheses used to generate data or to the data used to generate hypotheses). With a model-centered paradigm, the reproducibility focus changes from the ability of others to reproduce both data and specific inferences from a study to the ability to evaluate models as representation of reality. Validation underpins ‘natural selection’ in a knowledge base maintained by the scientific community. The community itself is thereby supported to be more productive in generating and critically evaluating theories that integrate wider, complex systems.
Similar content being viewed by others
Introduction
Hypothesizing after the results are known (HARK), a term coined by Kerr (1998), defines the presentation of a post hoc hypothesis as if it had been made a priori. HARK is often viewed as inconsistent with the hypothetico-deductive method, which articulates a hypothesis and an experiment to falsify or corroborate that hypothesis.
HARK can lead to data dredging, data fishing, or p-hacking that is unduly manipulating data collection or statistical analysis in order to produce a statistically significant result. Data dredging has been fervently debated over the past two decades (Browman and Skiftesvik, 2011; Mazzola and Deuling, 2013; Head et al., 2015; Lakens, 2015; Bin Abd Razak et al., 2016; Bosco et al., 2016; Bruns and Ioannidis, 2016; Hartgerink et al., 2016; Nissen et al., 2016; Amrhein et al., 2017; Hartgerink, 2017; Hollenbeck and Wright, 2017; Prior et al., 2017; Raj et al., 2017; Rubin, 2017; Wicherts, 2017; Hill et al., 2018; Turner, 2018; Wasserstein et al., 2019)
While the term HARK arose from epistemological debate, reflecting arguments from logical empiricism to falsification (Kerr, 1998; Wagenmakers et al., 2012; Bosco et al., 2016; Rubin, 2017), the negative connotations of HARK in publication bias and p-hacking has become its primary label (Simmons et al., 2011).
HARK has not been yet set against the modern reality of a duty to use big, ubiquitous, ‘organic’ data, integrated from multiple, diverse sources, e.g., from high-throughput genomic sequencing to social media streams. The organic nature of big data is customarily defined in terms of big Vs –as in volume, variety, velocity, veracity (Chartier, 2016)– typifying high dimensionality (enormous sample sizes and feature spaces), heterogeneity, dynamicity, and uncertainty. Big data, coupled with unprecedented computational power, can potentially be used to generate and test many hypotheses and models.
Big data expand and evolve. The organic property of big data can make a study both prospective and retrospective, fusing exploratory and hypothesis-focused research into a continuum that challenges methodology. This makes a classical hypothetico-deductive framework inadequate because deductions can lead to abductions as data collection proceeds. HARK becomes inevitable, but it could be made legitimate and contribute to a more complete interpretation of the big data’s (latent) signals. However, robust methodological safeguards must be undertaken to avoid data dredging and the threats to reproducibility from HARK.
The objective of the present work is to contextualize HARK in big data research and propose an operational framework toward reproducible inference and theories. The roadmap to this paper is as follows. We first summarize a consensus on tackling HARK and p-hacking within the hypothetico-deductive framework. Second, we explore how recommended practices to avoid HARK, e.g., preregistration, may not cope with big organic data. Third, we contextualize big data studies within a hybrid hypothetico-deductive and abductive theoretical paradigm, illustrating how HARK is unavoidable. Fourth, we outline a HARK-solid, operational framework of reproducible inference on big data that focuses on models (as formalization and empirical verification of hypotheses) and emphasizes multifaceted study design, many data realizations, and massive model testing, all underpinned with statistical rigor in modeling and multi-level validation. The term ‘solid’ does not mean that HARK is prevented from happening, but rather that its occurrence is transparent and part of the inference process. Finally, we discuss how the outlined framework can be implemented in practice and how it can aid the scientific community to develop deeper theories for complex phenomena.
Overview on HARK
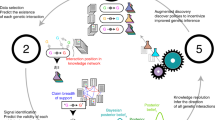
Figure 1 illustrates a typical HARK inference path: An initial hypothesis (A) is formed from real-world evidence or a standing mechanistic theory. A study to test the hypothesis is designed and data collected, but, after analyses, the hypothesis is weakly supported. The report of this experiment is rejected by journals who consider it unimportant. Through data dredging, e.g., addition of more data or changes on the model/covariates of interest, hypothesis (A) is confirmed, with potential p-hacking. Reflecting on the results, another hypothesis (B) is formed and supported by the first or second data sample, i.e., HARK happens. The full HARK process is rarely reported—just those activities directly related to the results in a given paper. The authors of the report have several options: (1) use original data, suppress hypothesis (A), substituting it with (B), and report results on (B); (2) report results on both (A) and (B) regardless, using the original data or the augmented data; (3) suppress results on (B) and report data dredged results on (A); (4) suppress everything, i.e., do not publish. All of the above options deviate from the hypothetico-deductive method and generate a number of potential threats to research validity, including publication bias.
HARK has been deconstructed in various ways (Rubin, 2017), including constructing, retrieving, and secretly suppressing hypotheses after results are known (CHARK, RHARK, SHARK). Not all types of HARK are considered detrimental: for instance, transparent HARK (THARK), i.e., reporting and discussing post hoc exploratory data analysis on empirical studies, has been deemed “beneficial to scientific progress and, in many cases, ethically required” (Hollenbeck and Wright, 2017). Nonetheless, it is often not possible to distinguish what kind of HARK has happened in a study.
HARK and data dredging can take place due to costs and rewards: the costs being associated with collecting more data and running more experiments to test multiple hypotheses or to redesign and rerun studies; and the rewards for publishing negative studies being relatively low. The recognition that a large proportion of published research is likely to be false through conscious or unconscious HARK and data dredging (Macleod et al., 2014; Begley and Ioannidis, 2015) has ignited a reproducibility crisis, especially in social, health, and biomedical sciences (Baker, 2016).
The scientific community reacted: Munafo et al. (2017) published “A manifesto for reproducible science”, to improve research efficiency and robustness of findings. Regarding the hypothetico-deductive framework, they noted a vicious circle of threats to reproducible science: generation of specific hypothesis (with failure to control for bias) → study design (low statistical power) → data collection and study conduction (poor quality control) → data analysis (p-hacking) → interpretation of results (p-hacking) → publication (selective bias). In this circle, HARK links results interpretation and generation of hypotheses.
The manifesto’s many resolutions included: more rigor in methodologies, defense from cognitive bias, promotion of study preregistration, protection against conflict of interest, encouragement of transparency, diversified peer review, upholding of standard research and reporting guidelines (Vandenbroucke, 2007; Little et al., 2009; Schulz et al., 2010; Nosek et al., 2015; Korevaar et al., 2016), and providing rewards for open and reproducible practices.
Among the manifesto’s calls, preregistration is central. Study preregistration is the publication of a hypothesis, design choice, and analytical approach before starting the data collection (Nosek et al., 2018). Preregistration seeks to eliminate shifts in evaluation of studies based on results and, therefore, it is intended to prevent cognitive bias of the study promoters (i.e., self-deception), publication bias (i.e., suppression of negative results), a number of HARK types (e.g., SHARK), and other data dredging. Registered reports (Chambers, 2013) are a form of preregistration that includes preliminary peer review. In brief, registered reports split the peer review process into two stages, before and after the study is carried out. A study design is thus evaluated before data are collected and analyzed; in addition, any post hoc analyses are easily recognizable as per the process, allowing flexibility in secondary analyses.
Despite the general consensus, there has been skepticism about both the theoretical validity and practical feasibility of preregistration (Lash and Vandenbroucke, 2012; Ioannidis, 2015; Vandenbroucke, 2015; Vancouver, 2018). Gelman and Loken (2013) epitomized the “garden of forking paths”: when there are always many choices for analyzing the data, the choice being more or less influenced by the modeler’s look at the data. They recommended that “the best strategy is to move toward an analysis of all the data rather than a focus on a single comparison or small set of comparisons” when preregistration is not an available option. In relation to registered reports, their implementation relies on the approval of an external review board, which is usually under a journal’s or the funder’s aegis; therefore, registered reports are prone to confirmation bias, and not free from potential conflict of interest.
The hypothetico-deductive framework works well with randomized controlled experiments, such as clinical trials, where confounding is controlled and other external conditions are made constant. The experiment is conceived to test just one or few, very specific hypotheses. In the narrow set of conditions where simple theory is appropriate, and the questions addressed are not highly enmeshed in complex systems, preregistration is advisable and should be included in research guidelines. However, when there is valuable data that could inform various inferences, such data should not be seen as inappropriate for scientific inferences just because it cannot be fit into an inference flow that is compatible with preregistration and registered reports. Notably, Munafo et al. (2017) and Nosek et al. (2018) specified that preregistration and registered reports do not apply to exploratory analyses.
We argue that the countermeasures proposed by Munafo et al. (2017) are not appropriate for a set of new situations that are becoming typical of big data science. These conditions imply not only big complex data, but big, complex, highly evolved, natural systems. In complex systems settings, exploratory analyses and hypothesis testing can work together, and therefore a new covenant on HARK is needed. The motivation behind our work originates from the recognition of the potential utility of HARK, as epitomized by Vancouver (2018). However, we are agnostic to assigning positive or negative connotations to HARK (or to CHARK, SHARK, THARK, etc.) and we stand by the neutral formalization provided by Gelman and Loken (2013). Gelman and Loken (2013), Munafo et al. (2017), Nosek et al. (2018), and others have previously discussed cases where preregistration may not be an option: we address in depth these scenarios. Lash and Vandenbroucke (2012) already argued that preregistration is unlikely to improve inference, affirming a flawed analogy of preregistering epidemiology and psychology studies to randomized controlled trials, and questioning definition consistency and practical feasibility of preregistration. They proposed instead to share openly data with important metadata information, such as the description of data generation and the prior uses. While open data and metadata are desirable and are becoming the de facto practice standard in research, they might not provide a sufficient condition to curb data dredging. In fact, preliminary evidence of issues with preregistration has been brought up by an empirical study (Claesen et al., 2019), which urges reflections and awareness on a stubbornly open problem.
Prediction and postdiction with big data
Technological progress has reduced the cost of generating or collecting data in many research fields. Big data raise research ambitions but come at the price of poorly harnessed heterogeneity, uncertainty, obsolescence, and many other hurdles that affect the process of study design and hypothesis testing or generation.
In the “Preregistration revolution”, Nosek et al. (2018) point out the utmost importance of distinguishing between prediction (testing hypotheses) and postdiction (generating hypotheses) because presenting postdiction as prediction is essentially HARK, and it threatens reproducibility. They advocate preregistration as the best mean to distinguish prediction from postdiction, assuring that “the problem of forking paths is avoided because the analytic pipeline is specified before observing the data”. Nonetheless, they acknowledge that challenging situations—gray areas—can occur; for instance, data collection procedures can change or be violated over the course of experiments, part of the data may be pre-existing, data can be dynamical, data can be multivariate, a glut of hypotheses could be preregistered, or hypotheses could be weak. In such cases, they claim that still preregistration ameliorates the likelihood of reporting false results; hence THARK is indulged and given clearance.
With big organic data, the aforementioned challenges abound. The boundaries between prediction and postdiction are blurred, as not all big data studies are exploratory, and HARK can be inevitable.
Therefore, for situations where the hypothesis space is large, or where there are many ways to generate data, the best practice guidance for hypothetico-deductive research needs to be relaxed or revisited in relation to other paradigms, such as abductive reasoning (Douven, 2013; Douven and Schupbach, 2015). Abductive reasoning, in the sense of inference to the best explanation, starts from the data and seeks to find the most likely hypothesis. A cyclic hypothetico-deductive and abductive epistemological framework was proposed by Ramoni et al. (1992) over 25 years ago—before the popularization of big data and data science—for knowledge based systems and, in particular, for medical knowledge. In their framework, prediction and postdiction cycle continuously: prediction follows the hypothetico-deductive process, and postdiction is abductive. All exploratory analyses are abductive in nature, and all hypothetico-deductive experiments start from postdiction, i.e., preliminary evidence suggesting one plausible hypothesis to be tested. By deduction, hypotheses generate new data and findings that, by abduction, refine the hypothesis space for deduction.
With big organic data, abduction and deduction are hybridized rather than being cyclic. The hybridization occurs because the generation of data from hypotheses and the generation of hypotheses from data are entangled, can happen at the same time, and be indistinguishable, as shown in Fig. 2. For instance, two datasets generated on the same population to test two distinct hypotheses (e.g., physical activity lowers blood pressure and physical activity increases testosterone) could be integrated at some point with another newly generated dataset to test a third hypothesis (physical activity and genetics determine depression).
Hybridized hypothetico-deductive and abductive inference. In the hypothetico-deductive framework (blue), a hypothesis guides the setup of an experiment and the generation of data through which the hypothesis is confirmed or falsified, generating knowledge; in the abductive framework (red), pre-existing data—could have been generated by a deduction process too—are used to test many hypotheses, the most plausible of which is verified through deduction or retained into knowledge; with big organic data (green), distinction between hypothesis-generated data and data-generated hypotheses is more difficult, as data can be pre-existing, contemporary, or prospective in relation to many concurrent hypotheses. Thus, data can lead to hypotheses and hypotheses can lead to data, with new data and new hypotheses adding both in dependence and independently from the inference processes
To better illustrate the blurred boundaries of prediction and postdiction, we further give three examples of research involving large data spaces, with both strong and weak hypotheses, and many options for analysis. All examples contain inevitable (and sometimes undisclosed) HARK, infeasible preregistration, and need for further safeguards against p-hacking and biased reporting.
Example 1: family genealogies
The evolutionary relationships and migration histories of organisms can be studied after genetic sequencing of species’ samples through molecular methods such as phylogenetics. A phylogenetic tree, similar to a family genealogy, is a binary tree whose leaves represent the sampled species, and the inner nodes represent ancestors from which the observed samples originated (Penny, 2004; Salemi et al., 2009). The leaves can also be annotated with dates, geographic locations, or other traits and estimate values for their ancestral states. A phylogenetic tree can, therefore, be used to test hypotheses of migrations like “The Cuban crocodile and other American crocodiles evolved from the Nile crocodile, which crossed the Atlantic, rather than from the Asian or Australian species” (Meredith et al., 2011) or “the Zika viruses found in the mosquito population in Florida originated from the Caribbean islands” (Grubaugh et al., 2017). In order to test the first hypothesis, evolutionary zoologists could collect samples from modern-day crocodiles or fossil samples, sequence their DNA, and verify. Similarly, for the second, epidemiologists would collect mosquitoes or human blood samples from different regions in Florida and other countries where Zika outbreaks have been reported. Nonetheless, even if a specific relationship or evolution hypothesis blooms in relation to preliminary evidence for solving a problem, phylogenetics is exploratory, post hoc in nature. This is because, given N genetic sequences from sampled species that have been aligned—this is already a process that involves high parameterization and uncertainty—there are (2N − 3)!! possible phylogenetic trees, i.e., evolutionary histories. The garden of forking paths is unmanageable already with 50 sequences (to say nothing of the evolutionary parameters in addition to the topology). Thirty years ago, only a few sequences could be obtained; nowadays, with advent of gene banks and high-throughput genetic sequencing, it is possible to collate different datasets with large sample size and longitudinal sampling, yielding hundreds, if not thousands, of isolates. The goodness of fit of a phylogenetic tree or of a tree branching pattern can be tested through different methods, and there are efficient heuristics that search for the best trees—in terms of likelihood or other optimality criterion—over such an enormous tree space. So, methods that can define the reduced tree space must be flexible and interpretable. Evaluating model robustness is essential, and methods such as likelihood ratio tests or bootstrapping are standard with many software applications. A predominantly Bayesian approach has emerged to handle the model complexity. Bayesian modeling helps dealing with the data uncertainty and the assumptions on the evolutionary models that are competing hypotheses themselves. Because the set of possible tree shapes is so huge, parameters constraining tree space are essential. Under a Bayesian framework the phylogenetic trees to be tested are assigned a prior probability based on evolutionary parameters. For instance, the rate at which genetic mutations appear in species can be modeled as a ‘molecular clock’, and different clock models, e.g., constant or exponential rates, can be tested. By numerically integrating over all possible trees, one can obtain marginal probabilities for hypotheses of interest. Extant evidence can be used as prior information for a new taxonomy search, and published evolutionary rates from other studies are often used to shape parameter sampling distributions.
Phylogenetics is not purely explorative; it makes assumptions about the effects of evolution and provides a framework for testing hypotheses. HARK occurs routinely, because it is intrinsic to the multi-hypothesis (tree topology) and multi-level (choice of models of evolution, model priors) nature of phylogenetic analyses. Sometimes such HARK is not completely transparent. HARK may be hidden by the choice of many alternative analysis pipelines that could be taken (e.g., maximum likelihood vs. minimum evolution tree search). Usually, data dredging is handled by robustness analysis, but it may still creep in when (1) the data collection is flawed, e.g., convenience sampling, or (2) when the wrong model set-up is employed. Rigor in methodology tackles (2), whilst (1) must be approached with proper study design and still relies on the initial hypothesis. Dataset collation in a phylogenetic study can lead to SHARK, as including or excluding a number of genome sequences can change the results in a more or less desired way, and therefore the published findings. Preregistration, however, is likely to be of little help here. One approach could be to declare which genome sequences are going to be used for analysis, given a strict rationale for excluding sequences, e.g., if they cannot be aligned or if they are too distant from the target species. In practice, these criteria may need to be very loose, since often decisions need to be made after analyzing data, making vain the purpose of preregistration itself. Better than preregistration, a consideration of existing scientific knowledge from literature could be embedded in the study design (merge existing and new data) or in the analysis as prior probability (narrowing the search space of evolutionary parameters already known). Moreover, as evolutionary theory becomes solidified by other studies, the entire phylogenetic analysis might deserve to be done completely from the start again under assumptions relevant to that new knowledge of evolutionary theory. The validity of a phylogenetic analysis is better assured by multiple investigators redoing an analysis given their understanding of the existing theoretical foundations for an analysis than it is by following some set of rules established in advance of an analysis.
Example 2: genomics and beyond
The human genome can be thought of as a string of a couple of billion letters (A, C, G, and T) representing nucleotides that hold the code for building and operating cells. On average, a person has about ten million variations, or single nucleotide polymorphism (SNPs), in their genome. Genome-wide association studies (GWAS) relate SNPs to (more or less) heritable health conditions and disorders (Manolio, 2010). A GWAS is usually considered as purely exploratory analysis. In fact, hypothesis generation in GWAS rarely focuses on the SNP. Rather, the focus is on gene function pathways or on macro evidence, such as a disease being observed at a high frequency in a particular subgroup (race, ethnicity, gender, socioeconomic status, etc.), e.g., “what are the genetic determinants of rheumatoid arthritis in Native American Pima, and how does their higher disease incidence compare to other ethnic/racial groups in North America?” (Williams et al., 1995). Classical GWAS typically involve a single dependent variable, a few million independent SNP variables, an ancestry genetic component, and other covariates. These million hypotheses are usually tested one-by-one using an allelic model with strict correction for multiple comparisons, meaning that the p-value shall be 5 × 10−8 or smaller. GWAS have been plagued with low statistical power since their inception, because the number of genotypes obtainable in a single study is constrained by technological limitations and costs. However, the landscape has changed rapidly with an explosion of GWAS in the past 20 years, including larger sample size, concurrent studies, standardized analytic pipelines, and meta-analyses. With costs of sequencing going down (in 2019, a mail order kit for SNP sequencing in the United States costs $99), it may be possible to sequence most people in high- and medium-income countries in the next few years, making the genotype a ubiquitous attribute.
Whereas it is straightforward to control the false discovery rate in GWAS, there may be other hurdles to replicability and reproducibility in hypothesis testing rather than in hypothesis generation. For instance, the definition of the disease phenotype can be arbitrary (say, a behavioral disorder), as well as the choice of a population structure model, or adjustment variables when testing associations with the phenotype. GWAS studies are generally regarded as replicable (Kraft et al., 2009; Heller et al., 2014; Rietveld et al., 2014), although there is variance among study phenotypes (Dumas-Mallet et al., 2016; Arango, 2017). The current “big short” with GWAS is in the missing heritability—i.e., why single genetic variations cannot account for much of the heritability of diseases—and the poor predictive performance of SNP-based risk scores, also due to non-hereditary factors (Marigorta et al., 2018). In other words, we know the genes that are associated to a disease phenotype, but we cannot predict accurately if a person is going to develop the disease based on their genes. Genomics can now be coupled with other omics domains, e.g., the transcriptome, the microbiome, the proteome. Each omics domain can add millions of additional variables. Consequently, a GWAS-like approach to find associations with phenotypes is very limiting. The purpose of multi-omics studies, after all, involve looking at cross-domain mechanisms. Incorporation of cross-domain mechanistic theory can improve prediction accuracy of phenotypes. Such a mechanistic theory might relate to which SNPs act in concert with different types of joint effects, such as simple independent action or multiplicative effects. Failure to formulate joint effects correctly might be one reason for the low predictive power of GWAS studies. The principles of formulating joint effect models described by epigenesis theory might help overcome this deficiency (Koopman and Weed, 1990). As biological theory advances, the theoretical basis of defining joint effects is becoming increasingly established. Without such theory, the severe computational burden associated to fitting joint probability parameters for large numbers of SNPs limits the number and the complexity of hypotheses that could be tested. Finally, with both GWAS and multi-omics studies, the cumbersomeness in data generation and collation makes it difficult to clearly distinguish prospective and observational designs. For instance, genomic sequencing of a population obtained for testing SNP associations with diabetes could also be used to test retinopathy. Genomic sequencing could also be obtained as routine screening of public health utility independently from an outcome of interest.
Riva et al. (2010) presented an operational framework for GWAS, based on the cyclic deductive-abductive model of Ramoni et al. (1992) which included refinement of phenotypes and integration with other knowledge base, implementing practically a full-fledged THARK. However, Riva’s framework is only in part exempt from threats to reproducibility; as it covers hypothesis multiplicity for phenotypes and basic validation, but not other forking paths such as model choice and reporting bias. For multi-omics studies, integrative approaches for study design have been proposed to recognize the “enduring availability [of large—omics data sets that] can be reanalyzed with multiple approaches over and over again” (Hasin et al., 2017). The analytical challenges of integrating omics data into GWAS analyses are considerable. Many approaches to generating hypotheses and theories arise. These might focus on the genome, the phenotype, or the environment. The rewards could be considerable, but such integration will use HARK to build the needed complex theoretical base.
Example 3: social media
Social media create streams of unstructured textual, image, audio and video data as well as structured (meta)data such as location. Some of the data are available at the level of individual users or populations (communities of interest, regions, organizations, etc.). The resolution of the data in time, place and person is high, uncovering new insights into human behavior. Through methods of social media data mining (e.g., natural language processing, network analysis, machine learning models for text/image categorization), the information can be progressively structured and used to answer research questions (Zafarani et al., 2014; Bian et al., 2019). A well-formed, preregistered hypothesis can be tested; an algorithm for data sampling can be shared, along with computer code to allow others to repeat the analyses on old or new data. Nonetheless, many forking paths in the data processing can threaten reproducibility. For example, consider a Twitter study that tests the hypothesis that “short-term and long-term non-contact martial arts training boosts work performance of middle-class employees in the United States”. The study design may include selection of tweets based on geolocation, citizenship, removal of spammers/bots, classification of job satisfaction (e.g., positive, negative, neutral), etc. The number of parameters involved in natural language processing or deep learning analysis is usually very high. The uncertainty of machine learners to categorize tweets can be also high. Even if the hypothesis and its testing procedure have been specified properly, the experimental data generation and encoding can be highly variable. In addition, large imbalance between cases and controls creates situations in which a relatively small-sample case set can be compared to at-a-will number of control groups. A few changes in the data collation and feature extraction/categorization pipeline could produce largely different datasets with the same study design. The massive variability in procedures and volatility of data can lead to uncontrollable HARK and all kinds of data dredging, severely affecting reproducibility. In a critical literature review of Twitter-based research on political crowd behavior, Cihon and Yasseri (2016) highlighted lack of standardized methods that permit interpretation beyond individual studies, remarking that “the literature fails to ground methodologies and results in social or political theory, divorcing empirical research from the theory needed to interpret it.” In another study, Pfeffer et al. (2018) illustrated artifacts in Twitter’s application programming interface, for which sampling should not regarded as random. They showed how it is possible to deliberately influence data collection and contents, consequently manipulating analyses and findings. Developing theory that handles such extreme variability is required, making HARK contribute to solidifying inferences rather than threatening their validity.
A HARK-solid inference framework for big data
In the context of big organic data, studies can clearly be affected by many types of HARK, and, as discussed, preregistration is not an efficient safeguard.
We, therefore, foresee a core theory based on the hybrid hypothetico-deductive and abductive paradigm—blending hypothesis testing and generation—that does not curb HARK but exploits its utility by decoupling it from data dredging, i.e., it makes it ‘solid.’ Such theory organizes a hypothesis’ space in relation to a data space and introduces multiple levels of hypothesis verification that form a model space.
We propose to shift the research focus from hypotheses to models. The difference is important. Hypotheses are so-called when they are yet to be formalized and tested—so to speak, they reside in Plato’s Hyperuranion. A model is a formalization of a hypothesis in the form of computable (over a defined input) function, multivariate joint probability distributions, or logical assert that can undergo empirical verification, i.e., Popperian falsification. Models are systematic, compressed representation of reality, and the model space is the gold standard to which one refers in both hypothesis generation and testing. Models therefore are the integration of hypotheses into an action-based structure. Models can capture the mechanistic aspects of processes that hypotheses may not. For instance, there are statistical models that focus on describing variation in relationships, machine learning methods for prediction or forecasting, and there are causal system models that focus on how different types of situations and events relate to each other in terms of causes and effects. Of note, models do not necessarily coincide with theory (or body of knowledge) because they are not always explicit or explanatory; for instance, a neural network model could provide a reliable representation of a complex physical phenomenon without being interpreted.
We recognize that the analytic workflow that leads to a model is tight to the model instantiation, and definitely aids with reproducibility. For this reason we are in favor of registration of research protocols or reporting guidelines rather than preregistration of hypotheses. However, we are more cautious in including the workflow component in the model definition because a model could be found though an irreproducible process, e.g., by intuition of a single scientist, and then survive falsification through multiple validations. Another term distinction we make here (not generally conventional) is between the terms method and model. Logistic regression or Bayesian phylodynamic tree estimation are methods. They become models when independent/dependent variables are defined, when coefficients/parameters are set or estimated, etc. In phylodynamics, a model is a finished tree (ensemble) with specified topology, branch lengths, node dates, locations, and species’ annotation.
Being model-centric is different from data-centric because the data become a mere falsification instrument and the knowledge (i.e., model) gains the prime role. To better illustrate how models are detached from hypotheses, and how models represent reality, in Fig. 3 we show a flowchart for knowledge discovery with big data, and a ‘natural selection’ framework based on internal and external validation that updates the set of existing models, i.e., the knowledge base of reality. The black links in the figure can be interpreted as conditional dependencies, like in a directed Bayesian network, while the blue links represent HARK and HARK-related phenomena. Note that here we are illustrating HARK that could be in service of legitimate new knowledge generation. In our scheme, all of the HARK-like links originate from models. They set off processes that can change conceptualizations of what is important in real-world evidence, how we should think about the problem we are addressing, what hypotheses are most important to explore, what our research objectives are, or how we need to design a study to advance knowledge. Without these HARK-like links, one vastly restricts the potential to generate new knowledge, as in the three case-study examples we have presented. Thus excessive rigidity in controlling HARK could restrict scientific advancement rather than eliminate non-reproducible research results.
A scheme for HARK-solid knowledge inference and model ensemble update. The orange compartments (real-world evidence and problem/research objective) represent the motivating forces for organizing a research endeavor. The gray compartments (hypotheses and study design) represent the intellectual effort to organize that research. The green compartments (available data/experiment sources, data sample and other data/experiments) represent the data one can use in pursuit of the inferences one seeks in regard to hypotheses or the construction of new models relevant to the real world. The purple components represent inference (new models) and knowledge bases (existing models). The magenta compartments (internal and external validation) authenticate inferences made by analyzing data with new or existing models: internal validation of inferences comes from study-generated models and data, while external validation comes from other data and models. The azure lines represent model generated actions that could lead to HARK. For the examples we have presented, this HARK is helpful for advancing science rather than generating non-reproducible results
We now explain Fig. 3 starting from the top node and following the direction of black arrows.
-
i.
Based on some real-world evidence or prior theoretical scrutiny, a problem or a research objective can be posed: for instance, in the 1950s, lung cancer became a top cause of death worldwide and scientists needed to find the cause and, possibly, a solution. A number of hypotheses to explain the problem springs from the evidence and from existing knowledge and theory; the latter can be prejudiced by a form of cognitive bias (e.g., confirmation bias).
-
ii.
A study design is generated around the hypotheses, which can be many in case of explorative analysis, where a general overarching hypothesis can basically contain many, such as in GWAS. Study design is necessarily influenced by extant hypotheses and theory manifest in existing models but also indirectly by available data/experiment sources. A randomized controlled trial is designed to minimize selection bias when testing the effect of a new treatment (the hypothesis). However, a randomized trial is not always feasible: for instance, it would not be ethical to design a trial to address whether smoking causes lung cancer by assigning subjects randomly to smoking. Thus, an observational study would be the ethical alternative, but observational studies are more prone to confounding and bias. Further, as discussed, both prospective and observational studies designed to test one hypothesis could be used to test many others. An implementation of a study design, e.g., a standard operating procedure for data extraction, can also generate additional variability due to the choice of sampling parameters (as we showed in the social media example).
-
iii.
Data samples are generated from available data/experiment sources and study design. A study design might draw from multiple data bases. For instance, a retrospective study to test the association between smoking and lung cancer, adjusted by genetic background, is designed. The study is implemented by querying medical records that document both smoking habits and genetic testing. The researchers identify two sites: one healthcare provider serves a large population, but its clinical information system allows queries only on past/current smoking and a certain number of genes; the other provider has more detailed behavioral data (including number of cigarettes per week) and higher-resolution genetics but serves fewer people and provides less data. Notably, the conditional dependency of hypotheses, study design and data samples is not necessarily conform to the hypothetico deductive paradigm, but can be abductive or hybrid—as many cases are with big data—because of the other conditional link coming from available data/experiment sources and of the multiple hypothesis context.
-
iv.
The study design and the data sample entail the inference of new models. Each model represents one of the many ways to test how hypotheses are supported by a data sample. For instance, following the lung cancer study design example, the model could be a logistic regression on the lung cancer outcome with smoking and SNPs as covariates. Nonetheless, various alternative models on the same data could be set up, and existing models can be tested too. The notion of model in this scheme is generic and spans from simple relationships between measured variables to more complex representations, static or dynamic; the new/existing models can be purposed/repurposed to discover association, perform predictions or explain causality. Note that a number of HARK links generate from here to hypotheses and study design (discussed below).
-
v.
Internal validation (e.g., cross-validation) is used to ensure parameter robustness and generalizability among the various model choices on the specific data sample; the goodness-of-fit is dependent on the research objective and the model purpose, e.g., an effect size, a prediction error or the strength of conditional independence.
-
vi.
The inferred new models then undergo a second level of verification by being compared with existing models (and/or model forms that differ from those in the original study design) through external validation, using datasets that have been generated independently from the starting hypotheses and from the study design that led to the data sample. To some extent, the external validation can be considered abductive as the models being tested represent hypotheses. For instance, the logistic regression model derived using smoking habits and SNPs could be compared with an existing genetic risk score on a new dataset collated from multiple sources of healthcare records. As another example, the relationships of interest could be analyzed using existing models that have different joint effects implications. Ultimately, new models update the existing models set, by joining the model space, rewiring the knowledge base and theory.
In our conceptual framework there are two main HARK-like paths. The first set of paths that arise from the new models node and lead to changes in the hypotheses or study design, which may subsequently change the data sample. If we assume that appropriate statistical procedures are used, internal validation can handle post hoc changes in hypotheses, using multiple model comparison, especially if existing models are factored in. However, if the changes in hypotheses and study design affect the data sample—generation of data biased by postdiction—then internal validation is not effective because it relies on the data sample itself. For instance, in a GWAS study on depression, the phenotype could be changed by using a different symptom scoring method, then used to identify more associated SNPs. External validation is necessary because other, independent data samples are not affected by changes in hypotheses or study design—the phenotype definition in the GWAS example. One could argue that the collation of other data is dependent on the study design because of sampling parameters, but the external validation does not need to be carried out within the same study and can be operated by independent researchers who decide to use a different design. In the end, validation of many models over many datasets results in ‘survival of the fittest’ model(s).
The other HARK-like paths arising from the existing models node represent both the cognitive bias and the effect that the models have in shaping our perception of the real world. For example, in the phylogenetic analysis cases outlined above, it could be difficult to accept alternative evolutionary histories that contradict longstanding evidence (even if derived from old, biased data and obsolete methods).
In summary, reproducibility is attained by maximizing the generalizability of models and minimizing the bias in model space (i.e., the bias as to what we perceive to be a valid model space). The latter is more problematic because it biases the data generation and therefore external validation.
The manifesto for reproducible big data science
We recommend the following actions to progress beyond preregistration toward HARK-solid approaches that can better handle reproducibility in big data inference. Our recommendations apply mostly to descriptive/prediction models, but they could be extended to interventional, causal models that rely on stronger assumptions, domain knowledge, and prospective design.
-
Exploit hypothesis multiplicity. While pure hypothetico-deductive studies, like clinical trials, are defined with specific hypotheses a priori and well handled by preregistration, with big data studies –as described– prediction and postdiction have blurred boundaries. Big dataset collation must therefore be transparent, i.e., agnostic, to the hypotheses of choice, because new hypotheses can emerge because of the continually growing, organic nature of big data. In these cases, enumeration of testable hypotheses as in “strong inference” may be pursued (Platt, 1964), together with data re-analyses when new variables or increased sample sizes are available, toward full-fledged abduction by explorative analysis. Hypothesis multiplicity may also help to reduce confirmation bias.
-
Focus on models, effect sizes and goodness of fit. A model can be as simple as the empirical confirmation of a single hypothesis in a controlled environment through a univariate test, a multivariate function, or a dynamic system representing a complex piece of reality (with or without causal meaning). Often, the effect sizes and the goodness of fit of a model (which can include functions usually used for assessing prediction performance, as well as methods for assessing causal plausibility) are neglected over the pursuit of statistical significance. With big data, where likely everything can be significant, we have a chance to revise the historical obsession on p-values, which leads to misconceptions and delusions about reproducibility (Gigerenzer, 1998; Wasserstein et al., 2019).
-
Exploit model multiplicity and perform internal validation. Model multiplicity involves both the set of factors/variables used and the statistical technique chosen. Nowadays, many statistical and machine learning techniques are available with methods for variable set selection. Statistical safeguards for internal (or in-sample) validation include more stringent p-value thresholds (Ioannidis, 2018) or type-2 error avoidance (Verhulst, 2016). For parameter optimization and model selection, robust techniques should be chosen, such as bootstrapping or cross-validation (Nadeau and Bengio, 2003; Hastie et al., 2009). When the parameter/model space is too large for grid searches or when cross-validation is resource-consuming (even if run in parallel), asymptotic selectors—quick to calculate—such as the Akaike, Bayesian, or other information criteria (Stine, 2004) can be used, accompanied by heuristic or random-like parameter/model searches, like simulated annealing. Stricter Bayesian formulations can be devised, assigning priors to models and selecting them based on a Markov Chain Monte-Carlo search and Bayes factors (Chipman et al., 2001; Faya et al., 2017). Also, multilevel modeling can be factored in (Gelman, 2006; Gelman et al., 2012). Internal validation and model multiplicity exploitation stand also in causal inference: for instance there are established approaches for automated learning of Bayesian networks and selection/evaluation of causal structures (Pearl, 2009). Still, there can be ill-posed scenarios where even internal validation can fail. Simonsohn (2014) showed through simulations (data peeking, convenient outlier removal, choosing favorable dependent variable or one variable combination out of many) how Bayes factors, not only p-values, can be hacked through selective reporting, where the only solution is “required disclosure.” Yet it could be argued that a stricter statistical approach to false discovery rate control or bootstrapping could ameliorate Simonsohn’s cases. Other improved inference strategies may include running variants of analytical pipelines in parallel, and adjusting false discovery rates accordingly (Carp, 2012; Heininga et al., 2015). There remain unanswered questions about optimal strategies, particularly in relation to uses of Bayes factors and common estimators of marginal likelihoods (Wolpert and Schmidler, 2012; van der Linden and Chryst, 2017).
-
Implement external validation. As we discussed, although internal cross-validation is usually robust, it is not sufficient for demonstrating generalizability of findings. External validation on other datasets, collected in different contexts, is required. There are well-established guidelines for external validation of association and prediction models, such as the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) (Moons et al., 2015; Colquhoun, 2017), as well as for causal inference, such as model transportability (Pearl and Bareinboim, 2014). Testing previously published findings and fitted models on new data (as distinct from meta-analysis of existing studies) is crucial to confirm and update the state-of-the art, i.e., the existing models available in literature and endorsed by the scientific community. Implementing proper external validation, however, may be not straightforward (Ulrich and Miller, 2015; Hartgerink et al., 2016; van Aert et al., 2016). Even meta-analyses used in disciplined evidence synthesis can be affected by publication bias (Sedgwick, 2015; Lin and Chu, 2018). Crowdsourcing analytics, which could be considered a form of concurrent internal/external validation, has been shown also to be highly unstable in cases of high peer rating (Silberzahn et al., 2018).
-
Ensure repeatability and replicability before reproducibility. We deconstruct the general reproducibility term into three aspects, according to Plesser et al. (2018). Study repeatability—defined as the possibility to reproduce results presented in a work with the same data—must be enforced in the peer review phases by promoting data and procedural/code/software sharing. Replicability and reproducibility are each important as: (i) the study design and/or the data collation procedure must generate similar datasets when applied in different settings or carried out by different teams, and consequently (ii) previous findings and/or inferences must be reproduced, i.e., confirmed. When repeatability and replicability are assured, e.g., code/software with possibility to explore preprocessing parameters, then internal/external validation assists toward reproducibility (or model generalization).
Potential pitfalls of the proposed approach
We have mentioned that one threat to our operational framework is the bias in model space, originating from multiple sources, including cognitive preconception and bias in external data generation. In particular, the requirement that models should be tested with data samples generated independently from the model/hypothesis generators may not be practical. In fact, whenever a validation occurs, someone has to design a data generation/collection and analysis workflow to test new and existing models. Indeed, the enormous space of parameters or design variations would apply to the buildup of this testing process. For example, if the testing dataset comes from Twitter, someone still has to decide how to remove spammers/bots, which tweets to select/filter, etc. In defense of the external validation we can note that it often comes from different research teams, and variations in the testing workflows could be seen as useful randomization to ameliorate unmeasured confounding bias. Yet, even when the researchers performing the test are not the same as those who built the model, we still cannot claim the design of the test to be fully independent from the model, given that the designers of the test must have known the model before figuring out how to test it. At present, we do not know if the variability in designs of external validation workflows is going to be enough to overcome such bias. A step in this direction might be to test one aspect of a model rather than the whole model, e.g., parameters, refocusing validation on to inferences rather than models. To a broader extent, one could validate the consistency of the data collation workflows.
Model and parameter validation bring us to a second problem of our framework, which is the definition of a validation methodology and model goodness criteria. For instance, when evaluating predictive models, a change in the performance function—entropy versus Brier score—can lead to different model rankings, affecting model selection in both internal and external validation. Even in simpler hypothesis testing scenarios, e.g., treatment is better than placebo, choosing a null hypothesis testing method instead of a Bayesian approach can lead to the of a multitude of results inconsistencies that flatten the model importance and hamper the survival of a subset of good models. In every study, the researchers may tweak the analyses to make sure that their model is better. There are many statistical validation methodologies which may be combined at will to favor one particular result. The plague of selective reporting based on p-values, and the misuse of p-values in general (Wasserstein et al., 2019) can still affect internal and external validation. To some extent, registered reports here may be resurrected because they allow the publication of negative results. Nevertheless, our proposed framework does not prevent, rather it emphasizes the publication of replication studies that validate someone else’s model, for which a more objective validation standard could be agreed by the relevant scientific community.
Future perspectives
Despite abundant literature on HARK, data dredging and p-hacking, there is little theory that can be used to test strategies for increasing reproducibility and decreasing publication bias. Preregistration and registered reports have become increasingly popular but need to demonstrate their effectiveness, since currently results are not clear (Allen and Mehler, 2018; Claesen et al., 2019). Our proposed framework for big organic data also requires assessment. A start could be to investigate how external validation affects survival of models in practice. For instance, one possible study could compare papers presenting prediction models (for a specific problem of interest) carried out following the TRIPOD guidelines (Collins et al., 2015) versus those that did not; the evaluation criteria could include common performance functions, citations or test in prospective populations. Another critical challenge that deserve further scrutiny are the effects of the variety of validation methodologies on the model space. Does the external validation converge on the true models, i.e., once one inspects a sufficient number of external datasets, does the fitness landscape of models reach an equilibrium?
Conclusions
We have summarized a spectrum of HARK-related reproducibility problems and opportunities, from narrow inferences made in the analysis of confined studies that can be p-hacked to big data explorations involving complex systems where HARK can become a virtue. That virtue of HARK expands the theoretical and inferential area being addressed. One dimension along the HARK vice-to-virtue scale relates simultaneously to the system complexity and to the distance of causal actions along causal chains from the measured variables to the measured outcomes being addressed. Those two scales are closely related because systems with extensive feedback and with complex patterns of joint effects between two variables generally have long causal chains. Even if causality is not the objective of the investigation, system complexity creates challenges for all types of modeling.
We presented a HARK-solid inference framework for big, ubiquitous and organic data, where prediction and postdiction often mingle, and commonplace safeguards do not work well. In hybrid abductive-deductive settings, HARK acquires a different epistemological role, becoming a building block of the inference process.
We have shifted the focus from hypotheses to models, which are (more computable) formalizations of hypotheses and provide their empirical verification. With a model-centered paradigm, the reproducibility focus changes from the ability of others to reproduce both data and specific statistical inferences from a study to the ability to evaluate representation of reality, theories, and the validity of scientific theory inferences. We highlighted the benefits—and limitations—of internal and external validation enabled by the data globalization, wherein inferred models are aired openly and are available for efficient falsification. One limitation of our framework is that it may be unfit for causal and dynamic system models. The models that could be most helpful for getting reproducible inferences from big organic data are causal process models, and the great advances in relating dynamic system models to data in recent years already call for new perspectives in analytics theory. Indeed, an interesting framework for inference in epidemiology that focuses on identifiability of parameters and causal theory misspecification, called inference robustness and identifiability analysis (IRIA), has been proposed by Koopman et al. (2016). IRIA may be considered almost HARK-solid, since it foresees model refinement through iterative data collection and robust inference, although does not explicitly addresses external validation.
Model validation through many organic datasets serves as a ‘natural selection’ criterion in the space of models/theories inferred and published by the scientific community. The community itself is facilitated to work more productively on generating and critically evaluating deeper theory that integrates more complex realities.
One could argue the futility of making inferences about hypotheses by examining data without an encompassing theory for the hypotheses being generated. Examining data should not be pursued for the purpose of evaluating hypotheses devoid of some larger theory encompassing the processes relevant to the hypotheses being considered. Without an overarching theoretical context, reproducible science cannot be assured. The reproducibility crisis has been often attributed to lack of transparency and statistical rigor, but a more deep-rooted problem is scientists’ “illusion of certainty” from established “statistical rituals” (Gigerenzer, 1998). Such rituals, like the focus on statistical significance, eliminate judgment and flatten scientific thinking. Science evolves through collaboration from diverse perspectives producing theoretical structures that wider groups build on. A HARK-solid framework for big organic data may help open and connect dialogue, theory and data research in a gestalt manner. The social change is that collaborative communities with highly diverse opinions coalesce around model inferences that make a difference either to policy or to science.
The cultural challenge may be greater than the theoretical or technical challenges in realizing HARK-solid frameworks. Cultures that force strongly held opinions to compete map more easily to Popperian ‘tearing down’ than to community-based, crowdsourced ‘building up’ of hypotheses, models and theory. If that community of scientists is not working to build a larger theoretical structure, if it just uses data to gather evidence for and against a single hypothesis, any methodological refinement for big data dredging will not contribute significantly to reproducible science.
Data availability
Data sharing not applicable as no datasets were generated or analyzed during this study.
References
van Aert RCM, Wicherts JM, van Assen MALM (2016) Conducting meta-analyses based on p values: Reservations and recommendations for applying p -uniform and p -curve. Perspect Psychological Sci 11(5):713–729. https://doi.org/10.1177/1745691616650874
Allen CPG, Mehler DMA (2018) Open science challenges, benefits and tips in early career and beyond. PLoS Biol https://doi.org/10.31234/osf.io/3czyt.
Amrhein V, Korner-Nievergelt F, Roth T (2017) The earth is flat (p > 0.05): significance thresholds and the crisis of unreplicable research. PeerJ 5:e3544. https://doi.org/10.7717/peerj.3544
Arango C (2017) Candidate gene associations studies in psychiatry: time to move forward. Eur Arch Psychiatry Clin Neurosci 267(1):1–2. https://doi.org/10.1007/s00406-016-0765-7
Baker M (2016) 1,500 scientists lift the lid on reproducibility. Nature 533(7604):452–454. https://doi.org/10.1038/533452a
Begley CG, Ioannidis JPA (2015) Reproducibility in science: improving the standard for basic and preclinical research. Circulation Res 116(1):116–126. https://doi.org/10.1161/CIRCRESAHA.114.303819
Bian J, Guo Y, He Z, Hu X (2019) Social web and health research: benefits, limitations, and best practices. https://doi.org/10.1007/978-3-030-14714-3. Accessed 27 Sep 2019
Bin Abd Razak HR, Ang J-GE, Attal H, Howe T-S, Allen JC (2016) P-hacking in orthopaedic literature: a twist to the tail. J Bone Jt Surg 98(20):e91. https://doi.org/10.2106/JBJS.16.00479
Bosco FA, Aguinis H, Field JG, Pierce CA, Dalton DR (2016) HARKing’s threat to organizational research: evidence from primary and meta-analytic sources. Pers Psychol 69(3):709–750. https://doi.org/10.1111/peps.12111
Browman H, Skiftesvik A (2011) Welfare of aquatic organisms: is there some faith-based HARKing going on here? Dis Aquat Org 94(3):255–257. https://doi.org/10.3354/dao02366
Bruns SB, Ioannidis JPA (2016) p-Curve and p-hacking in observational research. PLoS ONE 11(2):e0149144. https://doi.org/10.1371/journal.pone.0149144. Edited by D Marinazzo
Carp J (2012) The secret lives of experiments: methods reporting in the fMRI literature. NeuroImage 63(1):289–300. https://doi.org/10.1016/j.neuroimage.2012.07.004
Chambers CD (2013) Registered reports: a new publishing initiative at. Cortex Cortex 49(3):609–610. https://doi.org/10.1016/j.cortex.2012.12.016
Chartier T (2016) Vertigo over the seven V’s of big data. J Corp Account Financ 27(3):81–82. https://doi.org/10.1002/jcaf.22145
Chipman H, George EI, McCulloch RE (2001) The Practical Implementation of Bayesian Model Selection. In: Institute of Mathematical Statistics Lecture Notes-Monograph Series. Institute of Mathematical Statistics, Beachwood, pp 65–116
Claesen A, Gomes SLBT, Tuerlinckx F, Vanpaemel W (2019) Preregistration: comparing dream to reality. PsyArXiv. https://doi.org/10.31234/osf.io/d8wex, https://psyarxiv.com/d8wex/.
Collins GS, Reitsma JB, Altman DG, Moons KGM (2015) Transparent reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD Statement. Ann Intern Med 162(1):55. https://doi.org/10.7326/M14-0697
Colquhoun D (2017) The reproducibility of research and the misinterpretation of p -values. R Soc Open Sci 4(12):171085. https://doi.org/10.1098/rsos.171085
Cihon P, Yasseri T (2016) A Biased Review of Biases in Twitter Studies on Political Collective Action. Front Phys 4. https://doi.org/10.3389/fphy.2016.00034
Douven I (2013) Inference to the best explanation, dutch books, and inaccuracy minimisation. Philos Q 63(252):428–444. https://doi.org/10.1111/1467-9213.12032
Douven I, Schupbach JN (2015) Probabilistic alternatives to Bayesianism: the case of explanationism. Front Psychol 6. https://doi.org/10.3389/fpsyg.2015.00459
Dumas-Mallet E, Button K, Boraud T, Munafo M, Gonon F (2016) Replication validity of initial association studies: a comparison between psychiatry, neurology and four somatic diseases. PLoS ONE 11(6):e0158064. https://doi.org/10.1371/journal.pone.0158064. Edited by U S Tran
Faya P, Seaman JW, Stamey JD (2017) Bayesian assurance and sample size determination in the process validation life-cycle. J Biopharmaceutical Stat 27(1):159–174. https://doi.org/10.1080/10543406.2016.1148717
Gelman A (2006) Multilevel (hierarchical) modeling: what it can and cannot do. Technometrics 48(3):432–435. https://doi.org/10.1198/004017005000000661
Gelman A, Hill J, Yajima M (2012) Why we (usually) don’t have to worry about multiple comparisons. J Res Educ Effectiveness 5(2):189–211. https://doi.org/10.1080/19345747.2011.618213
Gelman A, Loken E (2013) The garden of forking paths: Why multiple comparisons can be a problem, even when there is no “fishing expedition” or “p-hacking” and the research hypothesis was posited ahead of time
Gigerenzer G (1998) We need statistical thinking, not statistical rituals. Behav Brain Sci 21(2):199–200. https://doi.org/10.1017/S0140525X98281167
Grubaugh ND, Ladner JT, Kraemer MUG, Dudas G, Tan AL, Gangavarapu K, Wiley MR, White S, Thézé J, Magnani DM, Prieto K, Reyes D, Bingham AM, Paul LM, Robles-Sikisaka R, Oliveira G, Pronty D, Barcellona CM, Metsky HC, Baniecki ML, Barnes KG, Chak B, Freije CA, Gladden-Young A, Gnirke A, Luo C, MacInnis B, Matranga CB, Park DJ, Qu J, Schaffner SF, Tomkins-Tinch C, West KL, Winnicki SM, Wohl S, Yozwiak NL, Quick J, Fauver JR, Khan K, Brent SE, Reiner Jr RC, Lichtenberger PN, Ricciardi MJ, Bailey VK, Watkins DI, Cone MR, Kopp IVEW, Hogan KN, Cannons AC, Jean R, Monaghan AJ, Garry RF, Loman NJ, Faria NR, Porcelli MC, Vasquez C, Nagle ER, Cummings DAT, Stanek D, Rambaut A, Sanchez-Lockhart M, Sabeti PC, Gillis LD, Michael SF, Bedford T, Pybus OG, Isern S, Palacios G, Andersen KG (2017) Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature 546:401
Hartgerink CHJ (2017) Reanalyzing Head et al. (2015): investigating the robustness of widespread p-hacking. PeerJ 5:e3068. https://doi.org/10.7717/peerj.3068
Hartgerink CHJ, van Aert RCM, Nuijten MB, Wicherts JM, van Assen MALM (2016) Distributions of p-values smaller than .05 in psychology: what is going on? PeerJ 4:e1935. https://doi.org/10.7717/peerj.1935
Hasin Y, Seldin M, Lusis A (2017) Multi-omics approaches to disease. Genome Biol 18(1):83. https://doi.org/10.1186/s13059-017-1215-1
Hastie T, Tibshirani R, Friedman JH (2009) The elements of statistical learning: data mining, inference, and prediction, 2nd edn. Springer (Springer series in statistics), New York
Head ML, Holman L, Lanfear R, Kahn AT, Jennions MD (2015) The extent and consequences of P-hacking in science. PLOS Biol 13(3):e1002106. https://doi.org/10.1371/journal.pbio.1002106
Heininga VE, Oldehinkel AJ, Veenstra R, Nederhof E (2015) I just ran a thousand analyses: benefits of multiple testing in understanding equivocal evidence on gene-environment interactions. PLoS One 10(5):e0125383. https://doi.org/10.1371/journal.pone.0125383. Edited by J Homberg
Heller R, Yaacoby S, Yekutieli D (2014) repfdr: A tool for replicability analysis for genome-wide association studies. Bioinformatics 30(20):2971–2972. https://doi.org/10.1093/bioinformatics/btu434
Hill MJ, Connell MT, Patounakis G (2018) Clinical trial registry alone is not adequate: on the perception of possible endpoint switching and P-hacking. Hum Reprod 33(2):341–342. https://doi.org/10.1093/humrep/dex359
Hollenbeck JR, Wright PM (2017) Harking, sharking, and tharking: making the case for post hoc analysis of scientific data. J Manag 43(1):5–18. https://doi.org/10.1177/0149206316679487
Ioannidis JPA (2015) Handling the fragile vase of scientific practices. Addiction 110(1):9–10. https://doi.org/10.1111/add.12720
Ioannidis JPA (2018) The proposal to lower P value thresholds to 0.005. JAMA 319(14):1429. https://doi.org/10.1001/jama.2018.1536
Kerr NL (1998) HARKing: hypothesizing after the results are known. Personal Soc Psychol Rev 2(3):196–217. https://doi.org/10.1207/s15327957pspr0203_4
Koopman JS, Weed DL (1990) Epigenesis theory: a mathematical model relating causal concepts of pathogenesis in individuals to disease patterns in populations. Am J Epidemiol 132(2):366–390. https://doi.org/10.1093/oxfordjournals.aje.a115666
Koopman J, Singh P, Iondies E (2016) Transmission Modeling To Enhance Surveillance System Function. In: MCNABB, S. (ed.) Transforming Public Health Surveillance: Proactive Measures for Prevention, Detection, and Response. Elsevier
Korevaar DA, Cohen JF, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, Moher D, de Vet HCW, Altman DG, Hooft L, Bossuyt PMM (2016) Updating standards for reporting diagnostic accuracy: the development of STARD 2015. Res Integr Peer Rev 1(1):7. https://doi.org/10.1186/s41073-016-0014-7
Kraft P, Zeggini E, Ioannidis JPA (2009) Replication in genome-wide association studies. Stat Sci 24(4):561–573. https://doi.org/10.1214/09-STS290
Lakens D (2015) Comment: what p-hacking really looks like: a comment on Masicampo and LaLande (2012). Q J Exp Psychol 68(4):829–832. https://doi.org/10.1080/17470218.2014.982664
Lash TL, Vandenbroucke JP (2012) Should preregistration of epidemiologic study protocols become compulsory?: reflections and a counterproposal. Epidemiology 23(2):184–188. https://doi.org/10.1097/EDE.0b013e318245c05b
Lin L, Chu H (2018) Quantifying publication bias in meta-analysis: quantifying publication bias. Biometrics 74(3):785–794. https://doi.org/10.1111/biom.12817
Little J, Higgins JPT, Ioannidis JPA, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, Scheet P, Gwinn M, Williamson RE, Zou GY, Hutchings K, Johnson CY, Tait V, Wiens M, Golding J, van Duijn C, McLaughlin J, Paterson A, Wells G, Fortier I, Freedman M, Zecevic M, King R, Infante-Rivard C, Stewart A, Birkett N (2009) STrengthening the REporting of Genetic Association Studies (STREGA)-an extension of the STROBE statement. Genet Epidemiol 33(7):581–598. https://doi.org/10.1002/gepi.20410
Macleod MR, Michie S, Roberts I, Dirnagl U, Chalmers I, Ioannidis JPA, Salman RA-S, Chan A-W, Glasziou P (2014) Biomedical research: increasing value, reducing waste. Lancet 383(9912):101–104. https://doi.org/10.1016/S0140-6736(13)62329-6
Manolio TA (2010) Genomewide association studies and assessment of the risk of disease. New Engl J Med 363(2):166–176. https://doi.org/10.1056/NEJMra0905980. Edited by W G Feero and A E Guttmacher
Marigorta UM, Rodríguez JA, Gibson G, Navarro A (2018) Replicability and prediction: lessons and challenges from GWAS. Trends Genet 34(7):504–517. https://doi.org/10.1016/j.tig.2018.03.005
Mazzola JJ, Deuling JK (2013) Forgetting what we learned as graduate students: HARKing and selective outcome reporting in I–O journal articles. Ind Organ Psychol 6(3):279–284. https://doi.org/10.1111/iops.12049
Meredith RW, Hekkala ER, Amato G, Gatesy J (2011) A phylogenetic hypothesis for Crocodylus (Crocodylia) based on mitochondrial DNA: evidence for a trans-Atlantic voyage from Africa to the New World. Mol Phylogenetics Evolution 60(1):183–191. https://doi.org/10.1016/j.ympev.2011.03.026
Moons KGM, Altman DG, Reitsma JB, Ioannidis JPA, Macaskill P, Steyerberg EW, Vickers AJ, Ransohoff DF, Collins GS (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 162(1):W1. https://doi.org/10.7326/M14-0698
Munafo MR, Nosek BA, Bishop DVM, Button KS, Chambers CD, Percie du Sert N, Simonsohn U, Wagenmakers E-J, Ware JJ and Ioannidis JPA (2017) A manifesto for reproducible science. Nat Hum Behav 1(1). https://doi.org/10.1038/s41562-016-0021
Nadeau C, Bengio Y (2003) Inference for the generalization error. Mach Learn 52(3):239–281. https://doi.org/10.1023/A:1024068626366
Nissen SB, Magidson T, Gross K, Bergstrom CT (2016) Publication bias and the canonization of false facts. eLife 5. https://doi.org/10.7554/eLife.21451
Nosek BA, Alter G, Banks GC, Borsboom D, Bowman SD, Breckler SJ, Buck S, Chambers CD, Chin G, Christensen G, Contestabile M, Dafoe A, Eich E, Freese J, Glennerster R, Goroff D, Green DP, Hesse B, Humphreys M, Ishiyama J, Karlan D, Kraut A, Lupia A, Mabry P, Madon T, Malhotra N, Mayo-Wilson E, McNutt M, Miguel E, Paluck EL, Simonsohn U, Soderberg C, Spellman BA, Turitto J, VandenBos G, Vazire S, Wagenmakers EJ, Wilson R, Yarkoni T (2015) Promoting an open research culture. Science 348(6242):1422–1425. https://doi.org/10.1126/science.aab2374
Nosek BA, Ebersole CR, DeHaven AC, Mellor DT (2018) The preregistration revolution. Proc Natl Acad Sci 115(11): 2600–2606. https://doi.org/10.1073/pnas.1708274114
Pearl J (2009) Causality: models, reasoning, and inference. Cambridge University Press, Cambridge, New York
Pearl J, Bareinboim E (2014) External validity: from do-calculus to transportability across populations. Stat Sci 29(4):579–595. https://doi.org/10.1214/14-STS486
Penny D (2004) Inferring phylogenies.—Joseph Felsenstein. 2003. Sinauer Associates, Sunderland, Massachusetts. Syst Biol 53(4):669–670. https://doi.org/10.1080/10635150490468530
Platt JR (1964) Strong inference: certain systematic methods of scientific thinking may produce much more rapid progress than others. Science 146(3642):347–353. https://doi.org/10.1126/science.146.3642.347
Plesser HE (2018) Reproducibility vs. Replicability: A Brief History of a Confused Terminology. Fron Neuroinform 11. https://doi.org/10.3389/fninf.2017.00076
Prior M, Hibberd R, Asemota N, Thornton J (2017) Inadvertent P-hacking among trials and systematic reviews of the effect of progestogens in pregnancy? A systematic review and meta-analysis. BJOG: Int J Obstet Gynaecol 124(7):1008–1015. https://doi.org/10.1111/1471-0528.14506
Raj AT, Patil S, Sarode S, Salameh Z (2017) P-hacking: a wake-up call for the scientific community. Sci Eng Ethics 24(6):1813–1814. https://doi.org/10.1007/s11948-017-9984-1
Ramoni M, Stefanelli M, Magnani L, Barosi G (1992) An epistemological framework for medical knowledge-based systems. IEEE T Syst man CY-S 22(6):1361–1375. https://doi.org/10.1109/21.199462
Rietveld CA, Conley D, Eriksson N, Esko T, Medland SE, Vinkhuyzen AAE, Yang J, Boardman Jason D, Chabris Christopher F, Dawes Christopher T, Domingue Benjamin W, Hinds David A, Johannesson M, Kiefer Amy K, Laibson D, Magnusson Patrik KE, Mountain Joanna L, Oskarsson S, Rostapshova O, Teumer A, Tung JY, Visscher PM, Benjamin DJ, Cesarini D, Koellinger PD, the Social Science Genetics Association Consortium, Eriksson N, Hinds DA, Kiefer AK, Mountain JL, Tung JY, Medland SE, Vinkhuyzen AAE, Yang J, Visscher PM, Conley D, Boardman JD, Dawes CT, Domingue BW, Rietveld CA, Benjamin DJ, Cesarini D, Koellinger PD, Conley D, Eriksson N, Esko T, Chabris CF, Johannesson M, Laibson D, Magnusson PKE, Oskarsson S, Rostapshova O, Teumer A, Visscher PM, Benjamin DJ, Cesarini D, Koellinger PD (2014) Replicability and robustness of genome-wide-association studies for behavioral traits. Psychological Sci 25(11):1975–1986. https://doi.org/10.1177/0956797614545132
Riva A, Nuzzo A, Stefanelli M, Bellazzi R (2010) An automated reasoning framework for translational research. J Biomed Inform 43(3):419–427. https://doi.org/10.1016/j.jbi.2009.11.005
Rubin M (2017) When does HARKing hurt? Identifying when different types of undisclosed post hoc hypothesizing harm scientific progress. Rev Gen Psychol 21(4):308–320. https://doi.org/10.1037/gpr0000128
Salemi M, Vandamme A-M, Lemey P (eds) (2009) The phylogenetic handbook: a practical approach to phylogenetic analysis and hypothesis testing, 2nd edn. Cambridge University Press, Cambridge, New York
Schulz KF, Altman DG, Moher D, for the CONSORT Group (2010) CONSORT 2010 Statement: updated guidelines for reporting parallel group randomised trials. BMJ 340(mar23 1):c332–c332. https://doi.org/10.1136/bmj.c332
Sedgwick P (2015) What is publication bias in a meta-analysis? BMJ h4419. https://doi.org/10.1136/bmj.h4419
Silberzahn R, Uhlmann EL, Martin DP, Anselmi P, Aust F, Awtrey E, Bahník š, Bai F, Bannard C, Bonnier E, Carlsson R, Cheung F, Christensen G, Clay R, Craig MA, Dalla Rosa A, Dam L, Evans MH, Flores Cervantes I, Fong N, Gamez-Djokic M, Glenz A, Gordon-McKeon S, Heaton TJ, Hederos K, Heene M, Hofelich Mohr AJ, Högden F, Hui K, Johannesson M, Kalodimos J, Kaszubowski E, Kennedy DM, Lei R, Lindsay TA, Liverani S, Madan CR, Molden D, Molleman E, Morey RD, Mulder LB, Nijstad BR, Pope NG, Pope B, Prenoveau JM, Rink F, Robusto E, Roderique H, Sandberg A, Schlüter E, Schönbrodt FD, Sherman MF, Sommer SA, Sotak K, Spain S, Spörlein C, Stafford T, Stefanutti L, Tauber S, Ullrich J, Vianello M, Wagenmakers E-J, Witkowiak M, Yoon S, Nosek BA (2018) Many analysts, one data set: making transparent how variations in analytic choices affect results. Adv Methods Pract Psychological Sci 1(3):337–356. https://doi.org/10.1177/2515245917747646
Simmons JP, Nelson LD, Simonsohn U (2011) False-positive psychology: undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychological Sci 22(11):1359–1366. https://doi.org/10.1177/0956797611417632
Simonsohn U (2014) Posterior-Hacking: Selective Reporting Invalidates Bayesian Results Also, SSRN Electronic Journal. https://doi.org/10.2139/ssrn.2374040
Stine RA (2004) Model selection using information theory and the MDL principle. Sociological Methods Res 33(2):230–260. https://doi.org/10.1177/0049124103262064
Turner DP (2018) P-hacking in headache research. Headache: J Head Face Pain 58(2):196–198. https://doi.org/10.1111/head.13257
Ulrich R, Miller J (2015) p-hacking by post hoc selection with multiple opportunities: detectability by skewness test?: Comment on Simonsohn, Nelson, and Simmons (2014). J Exp Psychol: Gen 144(6):1137–1145. https://doi.org/10.1037/xge0000086
van der Linden S, Chryst B (2017) No need for bayes factors: a fully bayesian evidence synthesis. Front Appl Math Stat 3. https://doi.org/10.3389/fams.2017.00012
Vancouver JB (2018) In defense of HARKing. Ind Organ Psychol 11(1):73–80. https://doi.org/10.1017/iop.2017.89
Vandenbroucke JP (2007) The making of STROBE. Epidemiology 18(6):797–799. https://doi.org/10.1097/EDE.0b013e318157725d
Vandenbroucke JP (2015) Preregistration: when shall we start the real discussion? Eur J Public Health 25(4):555–556. https://doi.org/10.1093/eurpub/ckv118
Verhulst B (2016) In defense of P values. AANA J 84(5):305–308
Wagenmakers E-J, Wetzels R, Borsboom D, van der Maas HLJ, Kievit RA (2012) An agenda for purely confirmatory research. Perspect Psychological Sci 7(6):632–638. https://doi.org/10.1177/1745691612463078
Wasserstein RL, Schirm AL, Lazar NA (2019) Moving to a world beyond “p < 0.05”. Am Statistician 73(sup1):1–19. https://doi.org/10.1080/00031305.2019.1583913
Wicherts J (2017) The weak spots in contemporary science (and how to fix them). Animals 7(12):90. https://doi.org/10.3390/ani7120090
Williams RC, Jacobsson LTH, Knowler WC, del Puente A, Kostyu D, McAuley JE, Bennett PH, Pettitt DJ (1995) Meta-analysis reveals association between most common class ii haplotype in full-heritage native americans and rheumatoid arthritis. Hum Immunol 42(1):90–94. https://doi.org/10.1016/0198-8859(94)00079-6
Wolpert RL, Schmidler SC (2012) α-Stable limit laws for harmonic mean estimators of marginal likelihoods. Statist Sin 22(3). https://doi.org/10.5705/ss.2010.221
Zafarani R, Abbasi MA, Liu H (2014) Social media mining: an introduction. Cambridge University Press, New York
Acknowledgements
We thank: Dr. Alberto Riva at University of Florida for the useful insights and hindsight on the cyclic abductive-deductive epistemological framework; Dr. Alessandro Vespignani at Northeastern University for pre-reviewing the paper and giving valuable feedback; Dr. George Michailidis and Dr. Marco Salemi at University of Florida for the useful discussions during the paper revision phases; finally, Dr. Brittany Rife at Temple University and Dr. Jae Sun Min at University of Florida for proof-editing the text and flagging obscure passages that needed clarification. This work was supported by US NIH-NCATS UL1TR001427 and UL1TR002389 grants, by US NSF SES 1734134 grant, by the University of Florida (UF) One Health Center, and by the UF “Creating the Healthiest Generation” Moonshot initiative, which is supported by the UF Office of the Provost, UF Office of Research, UF Health, UF College of Medicine and UF Clinical, and Translational Science Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Prosperi, M., Bian, J., Buchan, I.E. et al. Raiders of the lost HARK: a reproducible inference framework for big data science. Palgrave Commun 5, 125 (2019). https://doi.org/10.1057/s41599-019-0340-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1057/s41599-019-0340-8
This article is cited by
-
The benefits and pitfalls of machine learning for biomarker discovery
Cell and Tissue Research (2023)
-
Severe testing with high-dimensional omics data for enhancing biomedical scientific discovery
npj Systems Biology and Applications (2022)