Abstract
Abnormal mitochondria have been observed in bronchial- and alveolar epithelial cells of patients with chronic obstructive pulmonary disease (COPD). However, it is unknown if alterations in the molecular pathways regulating mitochondrial turnover (mitochondrial biogenesis vs mitophagy) are involved. Therefore, in this study, the abundance of key molecules controlling mitochondrial turnover were assessed in peripheral lung tissue from non-COPD patients (n = 6) and COPD patients (n = 11; GOLDII n = 4/11; GOLDIV n = 7/11) and in both undifferentiated and differentiated human primary bronchial epithelial cells (PBEC) from non-COPD patients and COPD patients (n = 4–7 patients/group). We observed significantly decreased transcript levels of key molecules controlling mitochondrial biogenesis (PPARGC1B, PPRC1, PPARD) in peripheral lung tissue from severe COPD patients. Interestingly, mRNA levels of the transcription factor TFAM (mitochondrial biogenesis) and BNIP3L (mitophagy) were increased in these patients. In general, these alterations were not recapitulated in undifferentiated and differentiated PBECs with the exception of decreased PPARGC1B expression in both PBEC models. Although these findings provide valuable insight in these pathways in bronchial epithelial cells and peripheral lung tissue of COPD patients, whether or not these alterations contribute to COPD pathogenesis, underlie changes in mitochondrial function or may represent compensatory mechanisms remains to be established.
Similar content being viewed by others
Introduction
Chronic obstructive pulmonary disease (COPD) is one of the leading causes of morbidity and mortality globally1,2. Classification of COPD disease severity is based on the degree of airway obstruction that results from pathological features in the bronchial compartment (chronic bronchitis) and the parenchyma (emphysema)3,4.
Inhalation of cigarette smoke (CS) and other noxious particles is the main risk factor for developing COPD5. After inhalation, toxicants and particles first reach the epithelial layer of the bronchial compartment where they can mediate damage and stress responses6. Changes in the composition and functionality of the bronchial epithelium in COPD patients (smokers) include basal- and goblet cell hyperplasia and aberrant number/length of ciliated cells7,8,9,10,11,12,13,14. During the development of COPD, however, not only the bronchial compartment but also the smaller airways and alveoli, are affected15,16.
In addition to structural- and functional changes described above, COPD is also characterized by aberrant cellular processes in epithelial cells of the airways and lungs including (chronic) inflammation cell death and oxidative stress16.
It has been shown that mitochondria, besides their role in energy metabolism, are critical regulators of these cellular processes17,18. Interestingly, abnormalities at the level of the mitochondrion have been described in COPD. Indeed, previous studies showed elongated and swollen mitochondria as well as fragmentation, branching and cristae depletion in primary bronchial epithelial cells (PBEC) from ex-smoking COPD patients with very severe disease compared to never-smoker controls19, and in PBECs from COPD patients relative to smokers without COPD20,21,22. Data on mitochondria in peripheral lung tissue of COPD vs non-COPD patients, likely due to the difficulty associated with obtaining parenchymal lung tissue, is almost non-existent. These changes are likely associated with exposure to CS as it has been convincingly shown that CS exposure of both bronchial-, small airway- and alveolar epithelial cells results in similar abnormalities in mitochondrial structure/phenotype19,20,23,24.
Mitochondria are dynamic organelles that are capable to adapt their morphology and energy production to meet demand. Mitochondrial homeostasis is preserved by the interplay between the synthesis of new organelles (mitochondrial biogenesis), and breakdown of damaged ones (mitophagy). Mitochondrial dynamics, controlled by fusion and fission events), is a key driver for these processes18,25.
Mitochondrial biogenesis is tightly regulated by transcriptional coactivators of the Peroxisome Proliferator-Activated Receptor (PPAR) Gamma (PPARG), Coactivator 1 (PPARGC1) family, respectively PPARGC1 Alpha (PPARGC1A), PPARGC1 Beta (PPARGC1B) and PPARG Coactivator-Related 1 (PPRC1), and their downstream transcription factors such as Nuclear Respiratory Factor 1 (NRF1), Nuclear Respiratory Factor 2 (NRF2) and Transcription Factor A, Mitochondrial (TFAM)26,27,28.
On the other side of the balance, mitochondrial quality control is mediated by the selective degradation of damaged or defective mitochondria by ubiquitin- or receptor-mediated mitophagy. Ubiquitin-mediated mitophagy is triggered by a loss of the mitochondrial membrane potential leading to stabilization and accumulation of PTEN-Induced Kinase 1 (PINK1) on the outer mitochondrial membrane. Subsequently, recruitment of Parkin RBR E3 Ubiquitin Protein Ligase (PRKN) to the mitochondria results in ubiquitination of mitochondrial proteins triggering the formation of an autophagosome and degradation of the organelle.
Receptor-mediated mitophagy is mediated by receptor proteins anchored on cytosolic side of the outer mitochondrial membrane, such as BCL2 Interacting Protein 3 (BNIP3), BNIP3-Like (BNIP3L) and FUN14 Domain Containing 1 (FUNDC1). These membrane receptors interact directly with Microtubule-Associated Protein 1 Light Chain 3 (MAP1LC3) Beta on the autophagosomal membrane, making ubiquitination redundant. In both mitophagy pathways, ultimately, specific general autophagy proteins are required to facilitate encapsulation of the damaged or defective mitochondria in an autophagosome29,30,31.
Cellular stress, as induced by for example inhaled CS, can result in alterations in mitochondrial bioenergetics and can induce mitochondrial dysfunction18,32,33,34.
Although previous studies have convincingly shown abnormal mitochondrial morphology/content in COPD, studies comprehensively assessing the molecular pathways controlling this are lacking. Moreover, although some studies have tried to address this in PBECs of COPD vs non-COPD patients, data on these pathways in peripheral lung tissue is almost non-existent.
Therefore, in the current study, our primary aim was to assess the abundance of essential regulators involved in mitochondrial turnover in lung tissue and cells of the airways from non-COPD patients and COPD patients at different stages of the disease. Biological material assessed in our analyses included peripheral lung tissue as well as isolated PBEC that were either cultured submerged (undifferentiated; basal cells) or at air–liquid interface (ALI) (differentiated; ciliated cells, secretory club cells, mucus-producing goblet cells). We hypothesized alterations in the abundance of key constituents associated with mitochondrial biogenesis, mitophagy, dynamics and metabolism in peripheral lung tissue as well as in bronchial epithelial cells from COPD patients compared to non-COPD patients.
Materials and methods
All methods were performed in accordance with the relevant guidelines and regulations.
Human peripheral lung tissue collection
Lung tissue was obtained from 7 patients with very severe emphysema (GOLDIV) who underwent lung volume reduction surgery, and had quit smoking for at least 6 months before the surgery. These patients were prescribed combination therapy of inhaled corticosteroids and long-acting β2-agonists, tiotropium/ipratropiumbromide and salbutamol, on demand. In addition, lung tissue was obtained from 6 control subjects with normal lung function (without cough and/or sputum production), and 4 moderate (GOLDII) COPD patients undergoing resection of a solitary peripheral tumor. For all subjects, a history of respiratory diseases other than COPD as well as increased respiratory complaints or respiratory tract infection during the 4 weeks preceding the study were considered as criteria for exclusion. The study was approved by the medical ethical committee of the Maastricht University Medical Center+ (MUMC+) (MEC 02-043.3). All subjects gave their informed consent in writing. Patient characteristics and clinical information are shown in Table 1.
Undifferentiated PBEC: collection, isolation and proliferation
Lung tissue used for the isolation of PBEC was obtained from the Maastricht Pathology Tissue Collection (MPTC). Collection, storage and use of tissue and patient data were performed in agreement with the “Code for Proper Secondary Use of Human Tissue in the Netherlands” (http://www.fmwv.nl). The scientific board of the MPTC approved the use of materials for this study under MPTC2010-019. In addition, the local medical ethical committee approved the use of human tissue (METC 2017-0087).
Isolation, culture and characterization of cells was performed by the Primary Lung Culture (PLUC) facility at the MUMC+ as previously described35,36. The PLUC facility provided PBEC isolated from resected bronchus rings remote from the diseased areas of patients undergoing lung surgery due to lung cancer at MUMC+ . PBEC from 7 donors without known history of COPD and 4 donors with known history of COPD (GOLDII) were provided for this study. Characteristics of the subject are summarized in Table S1.
PBEC of 11 donors (5 × 105 cells, passage 1) were thawed, seeded and expanded in T75-flasks (passage 2; Corning Costar) on pre-coated growth areas (coating: 10 μg/mL human fibronectin, 30 μg/ml PureCol Type I Collagen Solution (Bovine) and 10 μg/mL bovine serum albumin (BSA) diluted in Hank’s Balanced Salt Solution (HBSS; no calcium, no magnesium, no phenol red) (Gibco)) in Lonza Bronchial Epithelial Basal Medium supplemented with Bronchial Epithelial Cell Growth Medium (BEGM) singlequots (except Gentamycin) (Lonza), BSA (1.5 µg/mL) and 1% penicillin/streptomycin (Gibco) (i.e., BEGM complete). After medium refreshment every other day, confluence was reached upon 5–6 days of proliferation. Thereafter, PBEC were trypsinized, counted and seeded in quintuplicate in pre-coated 6 wells, 12 wells or 60 mm dishes in BEGM complete medium. Medium was refreshed every other day. Upon 85% confluence, PBEC were starved overnight using Lonza Bronchial Epithelial Basal Medium supplemented with Bronchial Epithelial Cell Growth Medium singlequots (except gentamycin, bovine pituitary extract and epidermal growth factor) (Lonza), BSA (1.5 µg/mL) and 1% penicillin/streptomycin (Gibco) (i.e., BEGM starvation). The next day, after 24 h of starvation, undifferentiated PBEC were harvested in appropriate buffers for follow-up analysis.
Differentiated PBEC: collection, isolation, proliferation and differentiation
PBEC from COPD patients (n = 6) and control subjects (n = 6) were collected at the University Medical Center Marburg in agreement with local ethics regulations (Marburg 223/12) (code 223/12). Briefly, bronchoscopies were conducted according to American Thoracic Society consensus procedure. During bronchoscopy, bronchial brushings were collected using protected specimen brushes. Cells were scraped from the brush and incubated in 8 mL accutase with gentle agitation for 2 min before 8 min centrifugation at 300 rcf. Cells were resuspended in Airway Epithelial Cell Growth Medium (AEGM, Promocell) and seeded at 0.5 − 1 × 105 cells/cm2. After passaging once by trypsinization, cryostocks (2 × 106 cells/vial) were prepared upon 80% confluence (passage 2). Well differentiated PBEC cultures at ALI were generated by culturing passage 4 cells on 6.5 or 12 mm transwell inserts (Costar 3460) for 28 days in 1:1 AEGM/DMEM (Gibco) supplemented with 0.1 ng/mL retinoic acid as described previously37. Barrier formation was monitored by measuring electrical resistance with an EVOM2 and STX2 chopstick electrodes. During measurements, the basolateral compartment was filled with AEGM:DMEM, and the apical compartment with Phosphate-Buffered Saline. To calculate transepithelial electrical resistance (TEER) in Ω*cm2, the resistance of a blank transwell was subtracted from the resistance of the cell layer, followed by multiplication with the transwell surface area. Characteristics of the COPD and non-COPD donors are summarized in Table S1.
RNA isolation and synthesis of cDNA
Human peripheral lung tissue
Total RNA was isolated from crushed human lung tissue using the Qiagen RNeasy kit (Qiagen, USA) according to the manufacturer’s instructions. Following analysis of the quantity and quality of the total RNA by nanodrop, total RNA (400 ng) was reverse transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Switzerland) following the instructions, including a no reverse transcription control and a no template control. Quantity and quality of the total RNA was verified by nanodrop analysis. Subsequently, cDNA was diluted in MilliQ (1:25) and stored in – 20 °C until use.
Undifferentiated PBEC
Total RNA was extracted from the undifferentiated PBEC (triplicates/donor) following the manufacturer’s protocol of the mirVana™ kit (Thermo Fisher Scientific, USA/NL). Following analyzing the quantity and quality of the total RNA by nanodrop, total RNA (800 ng) was reverse transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche) following the instructions. Subsequently, cDNA was diluted in MilliQ (1:25) and stored in – 20 °C until use.
Differentiated PBEC
Differentiated PBEC (duplicates/donor) were lysed in TRIzol™ Reagent (Invitrogen™) at day 0 (day of air-lift; undifferentiated) and day 28 (28 days after air-lift; fully differentiated). Total RNA was extracted according to the manufacturer’s instructions (Catalog number 15596026 and 15596018, Invitrogen™) using glycogen blue co-precipitant (Thermo Fisher Scientific, USA). Nanodrop analysis of the RNA was conducted to verify quantity and quality of the total RNA. Next, cDNA synthesis was conducted from 350 ng total RNA using iScript™ cDNA synthesis kit (Bio-Rad, the Netherlands) including a no reverse transcription control and a no template control. cDNA dilutions were made in MilliQ (1:50) and stored at − 20 °C until use.
Real time quantitative PCR analysis
Real-time quantitative PCR amplification was conducted by pipetting 4.4 µL of cDNA (diluted 1:25 or 1:50 in MilliQ water), 5 µL 2xSensiMix™ SYBR® and Fluorescein Kit (Bioline, the Netherlands) and 0.6 µL forward and reverse primers for target genes in white LightCycler480 384 multiwell plates (Roche). Next, to study gene expression, these samples were analysed on the LightCycler480 machine (Roche) following the thermal cycling protocol: 10 min at 95 °C, 55 cycles of 10 s at 95 °C, 20 s at 60 °C. The list of target-specific primers used for qPCR analysis is shown in Table S2.
Qualitative analysis of melt curves and peaks using LightCycler480 software (Roche) was used to check for a-specific amplification which were subsequently excluded. Quantitative analysis (i.e., N-zero values: extrapolated relative amount of DNA at cycle zero) by LinRegPCR software 2014.x (the Netherlands) was used to calculate gene expression. Exclusion of samples with no amplification, no plateau phase, too low Cq value or outside 5% of group mean was automatically conducted during LinRegPCR analysis. GeNorm software 3.4 (Primerdesign, USA) was used to calculate the geometric mean of a combination of at least two and up to four housekeeping genes (Actin B (ACTB), Beta-2 Microglobulin (B2M), Peptidylprolyl Isomerase A (PPIA), Ribosomal Protein L13A (RPL13A)), which was used for normalization of the calculated expression of target genes.
Western blotting
Human peripheral lung tissue
Crushed human lung tissue was lysed in buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, 0.1% NP40 and protease inhibitor cocktail (Roche).
Undifferentiated PBEC
Undifferentiated PBEC (quintuplicates/donor) were lysed in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Nonidet P40 or 1% Triton X-100, 0.5% NA deoxycholate, 0.1% sodium dodecyl sulphate in MilliQ) including fresh non-stable components: PIC, 1 mM sodium orthovanadate.
Differentiated PBEC
Differentiated PBEC (duplicates/donor) were lysed at day 28 in Whole Cell Lysis buffer (20 mM Tris pH 7.4, 150 mM NaCl, 1% Nonidet P40 in MilliQ) including PhosSTOP Phosphatase and cOmplete, Mini, EDTA-free Protease Inhibitor cocktail tablets (both Roche).
Whole cell lysates of human peripheral lung tissue, undifferentiated and differentiated PBEC experiments were rotated for 30 min at 4 °C and centrifuged at 20,000 × g for 30 min at 4 °C. Thereafter, the total protein content was analysed by the Pierce™ BCA Protein Assay Kit (Catalog number 23225 and 23227, Thermo Fisher Scientific), and whole cell lysates were diluted in a final concentration of 0.19–0.71 µg/µL in 1 × Laemmli buffer (0.25 M Tris pH 6.8, 8% (w/v) sodium dodecyl sulphate, 40% (v/v) glycerol, 0.4 M Dithiothreitol, 0.02% (w/v) Bromophenol Blue). These diluted lysates were boiled for 5 min at 100 °C and subsequently stored at − 20 °C (undifferentiated PBEC) or − 80 °C (lung homogenates/differentiated PBEC) until further analysis. The protein ladders (Precision Plus Protein™ All Blue Standards #161-0373, Bio-Rad) and protein samples were loaded on a Criterion XT Precast 4–12% or 12% Bis–Tris gel (Bio-Rad), respectively singular peripheral lung homogenates per donor (7.14 µg/sample), pool of quintuplicates per undifferentiated PBEC donor (10 µg/sample), and duplicates per differentiated PBEC donor (2.75 µg/sample). Proteins were separated in 1 × MES running buffer (Bio-Rad) by electrophoresis (100–130 V for 1 h), whereafter proteins were transferred from the gel to a 0.45 µM Nitrocellulose Transfer membrane (Bio-Rad) by electroblotting (100 V for 1 h) using a Bio-Rad Criterion Blotter. To stain and correct for total protein content, the nitrocellulose membranes were incubated with Ponceau S staining (0.2% in 1% acetic acid; Sigma-Aldrich) which was visualized using the Amersham™ Imager 600 (GE Healthcare, the Netherlands). To stain the target-specific proteins, the Ponceau S staining was washed off and the membranes were blocked for 1 h in 3% (w/v) non-fat dry milk (Campina, the Netherlands) dissolved in Tween20 Tris-buffered saline (TBST; 20 mM Tris, 137 mM NaCl, 0.1% (v/v) Tween20, pH 7.6). After finishing blocking, the membranes were washed with TBST and incubated with a target-specific primary antibody diluted in 3% (w/v) BSA at 4 °C overnight. Next, the membranes were again washed with TBST and now incubated with a horseradish peroxidase-conjugated secondary antibody diluted in 3% (w/v) non-fat dry milk in TBST for 1 h at room temperature. To visualize the target proteins, the membranes were washed with TBST, stained with 0.25-1 × Supersignal West FEMTO and 0.5-1 × Supersignal West PICO Chemiluminescent Substrate (Thermo Fisher Scientific), and imaged using the Amersham™ Imager 600. The list of primary and secondary antibodies used for western blotting is shown in Table S3. The Image Quant software (GE Healthcare) was used for quantification of the total protein content and target-specific proteins. Total protein content was quantified using the Ponceau S-stained images over the entire size range of proteins (250 kDa-10 kDa). This total protein content was used as normalization factor for (quantification of) target-specific proteins. Because several proteins of interest were detected on a similar gel, which is possible due to their difference in molecular mass, correction for total protein content/loading of these target proteins was based on the same Ponceau S staining. To specify, Gel I: HK2, DNM1L, OXPHOS, SQSTM1, TOMM20; Gel II: PRKN, BNIP3L, BNIP3, FUNDC1, PINK1, MAP1LC3B; Gel III: NRF1, TFAM, GABARAPL1, ESRRA.
Image acquisition: equipment and settings
We performed chemiluminescence capturing with automatic or manual exposure time as described in the Operating Instructions of the Amersham™ Imager 600 (GE Healthcare). A short pre-exposure is performed to determine the signal intensity. The system will use this information to calculate which exposure time will give the highest possible signal below saturation to enable accurate quantification of the sample.’ Manual exposure is only performed with a maximum duration of 5 min, in case the intensity of the image is inadequate (i.e., takes > 5–10 min) after automatic exposure, which is also suggested by the manufacturer’ instructions. Western blot images displayed in the (supplementary) figures of this manuscript have been adjusted for brightness and contrast equally throughout the picture. In addition, the Ponceau S band (37, 50, 70 or 75 kDa) presented in the (supplementary) figures is representative for the total Ponceau S staining per sample. For illustration purposes of the protein abundance in the human peripheral lung tissue homogenates in the manuscript figures, examples of bands of the target proteins along with the corresponding normalization bands of the Ponceau S staining are shown of one patient/group, which is not always representative for the mean and changes in the whole patient group as quantified in the box plots (due to the variation between patients). In the Supplementary Figs.1–3, all western blot images of the target proteins along with the corresponding normalization bands of the Ponceau S staining of all subjects/group are shown of the peripheral lung tissue from non-COPD patients and COPD patients (GOLDII; GOLDIV) (single) (Figure S1), as well as images of undifferentiated PBEC (pool of quintuplicates) (Figure S2) and differentiated PBEC (duplicates) (Figure S3) from non-COPD patients and COPD patients.
Statistical data analysis
GraphPad Prism 8.0 software (La Jolla, USA) was used to display the data in box plot or scatter plot graphs. Data are presented as mean fold change of COPD patients (GOLDII or GOLDIV) relative to non-COPD patients (box plot: min to max; scatter plot: ± standard error of the mean (SEM)). Individual subjects are represented by open circles (non-COPD patients), squares (all COPD patients), triangles (COPD GOLDII patients) and diamonds (COPD GOLDIV patients). Quality of the mRNA data was analysed by melt curve/peak analysis using LightCycler480 software (Roche). In addition, exclusion of extreme outliers in the analysis of peripheral lung tissue was also based on box plot analysis (i.e., values more than three interquartile ranges from the end of a box) in IBM Statistics SPSS 25. Statistical differences were analysed using GraphPad Prism 8.0 software. First, normal distribution of the data was assessed with the Shapiro–Wilk test. Secondly, in case of testing the difference between two groups (COPD patients versus non-COPD patients), a two-tailed unpaired parametric t-test (normal distribution) or non-parametric Mann–Whitney test (non-normal distribution) was used. In addition, in case of testing the difference between three groups (COPD GOLDII patients or COPD GOLDIV patients relative to non-COPD patients), we used a one-way ANOVA and a Dunnett’s post-hoc test for multiple comparisons (normal distribution) or a Kruskal–Wallis test followed by a Dunn’s multiple comparisons test (non-normal distribution). Moreover, the correlation between lung function (forced expiratory volume in first second/forced vital capacity; FEV1/FVC) and transcript levels of investigated markers was evaluated by calculating the Spearman’s correlation coefficient (ρ) using IBM SPSS Statistics version 25.0.
With respect to the validation of the PBEC differentiation, TEER and gene expression data are presented as mean fold change of day 0 versus day 28 per patient group. Statistical differences between day 28 (differentiated) relative to day 0 (undifferentiated) in separate patient groups were tested using a two-tailed paired parametric t-test, or in case of non-normal distribution a paired nonparametric Wilcoxon test. Moreover, differences between day 28 (differentiated) of non-COPD relative to COPD patients were tested using a two-tailed unpaired parametric t-test (normal distribution) or non-parametric Mann–Whitney test (non-normal distribution). Statistical significance was considered if p-values were below 0.05 (*p < 0.05) or 0.01 (**p < 0.01) and a trend was indicated if below 0.1 (#p < 0.1).
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. Human peripheral lung tissue: The study was approved by the medical ethical committee of the MUMC + , Maastricht, the Netherlands (MEC 02-043.3). All subjects gave their informed consent in writing. Undifferentiated PBEC: The scientific board of the MPTC approved the use of materials for this study under MPTC2010-019. In addition, the local medical ethical committee of the MUMC+ , Maastricht, the Netherlands approved the use of human tissue (METC 2017-0087). Differentiated PBEC: The study was approved by the local ethical committee Ethik-Kommission des Fachbereichs Humanmedizin der Philipps-Universität Marburg, Marburg, Germany (Marburg 223/12).
Results
Characteristics of COPD patients and non-COPD patients
Peripheral lung tissue was retrieved from non-COPD patients and COPD patients, of which the characteristics are described in Table 1. Obviously, lung function was significantly lower in the group of COPD patients (FEV1/FVC: 39.0 ± 5.1) versus non-COPD patients (FEV1/FVC: 83.1 ± 2.8). When COPD patients were categorized according to disease status (moderate: GOLDII and very severe: GOLDIV), the group of moderate COPD patients included only current-smoking males of an older age (69.8 ± 4.2) while the very severe COPD disease group consisted of mostly ex-smoking females (one ex-smoking male) of a younger age (53.3 ± 3.0). The differences in smoking status between these groups can be explained by the reason for surgery. Non-COPD patients and COPD GOLDII patients underwent surgery to remove a lung tumor and COPD GOLDIV patients underwent lung volume reduction surgery which requires smoking cessation. Body Mass Index (BMI) and pack-years were equally distributed among the groups.
Characteristics of non-COPD patients and COPD patients of which PBECs were cultured submerged (undifferentiated) or at ALI (differentiated) are summarized in Table S1. As expected, lung function was significantly lower in the group of COPD patients in both undifferentiated PBEC (FEV1/FVC: COPD: 60.2 ± 2.3 versus non-COPD: 79.3 ± 3.4) and differentiated PBEC (FEV1/FVC: COPD: 50.8 ± 4.0 versus non-COPD: 77.3 ± 2.0). In general, gender, age and BMI were equally distributed among the COPD and non-COPD patients in both PBEC studies. The only exception, COPD patients in the undifferentiated PBEC and both patient groups in the differentiated PBEC study included only males. Remarkably, pack-years was significantly higher in the group of COPD patients in differentiated PBEC (43.8 ± 5.7) versus non-COPD patients (7.5 ± 4.0).
Expression of epithelial cell markers in differentiated PBEC from COPD patients and non-COPD patients
Validation and characterization of the differentiation of PBEC was conducted by TEER (as an indication of the integrity of the formation of the epithelial barrier during differentiation) and by gene expression analysis of cell type-specific markers of basal and luminal epithelial cells on day 0 (undifferentiated) versus day 28 (fully differentiated).
As depicted in Figure S4A, differentiated PBEC from both COPD patients and non-COPD patients showed a significant increase in TEER from day 0 to day 28, while no differences were shown in TEER at day 28 between COPD and non-COPD patients. Moreover, as shown in Figures S4B and S4C, we observed a significant decrease in mRNA levels of basal epithelial cell markers (Tumor Protein P63 (TP63), Keratin-5 (KRT5) and Keratin-14 (KRT14)) as well as a profound increased expression of club cell secretory marker (Secretoglobin Familiy 1A Member 1 (SCGB1A1)), ciliogenesis marker (Forkhead Box J1 (FOXJ1)) and goblet cell differentiation marker (Mucin 5AC, Oligomeric Mucus/Gel-Forming (MUC5AC)) at day 28 compared to day 0 of differentiation in both non-COPD patients and COPD patients. Lastly, in COPD patients versus non-COPD patients, we observed that transcript levels of genes associated with club cell differentiation and ciliogenesis were decreased, while increased levels of a marker of goblet cell differentiation were observed at day 28 (Figure S4B,C).
Downregulation of key constituents controlling mitochondrial biogenesis in peripheral lung tissue from very severe COPD patients
To evaluate if, and to what extent, the pathways regulating mitochondrial turnover (biogenesis versus breakdown of mitochondria) are affected in peripheral lung tissue from COPD patients, we first investigated the mRNA expression of molecules involved in the regulation of mitochondrial biogenesis.
Peripheral lung tissue from COPD patients showed decreased mRNA levels of PPARGC1B and PPRC1 compared to non-COPD patients (Fig. 1A). Interestingly, upon further inspection, a significant downregulation of PPARGC1B and PPRC1 transcript levels was shown in COPD GOLDIV patients compared to non-COPD patients (Fig. 1B). Moreover, a significant positive correlation between lung function, i.e., percentage FEV1/FVC, and PPARGC1B expression (ρ = 0.609, p = 0.021; R2 = 0.371) as well as between lung function and PPRC1 expression (ρ = 0.613, p = 0.020; R2 = 0.397) was observed (Figs. 1C,D).
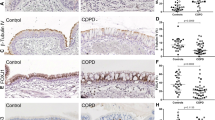
Decreased abundance of molecules controlling mitochondrial biogenesis in peripheral lung tissue from severe COPD patients. Real-time qPCR analysis was used to analyse mRNA expression of PPARGC1B and PPRC1 in human peripheral lung tissue from non-COPD patients (n = 5) and COPD patients (n = 9; GOLDII n = 4/9, GOLDIV n = 5/9) (A, B). Data are presented as mean fold change of COPD patients (GOLDII or GOLDIV) relative to non-COPD patients in box plots (min to max). Statistical differences between COPD patients versus non-COPD patients (two groups) were tested using a two-tailed unpaired parametric t-test (normal distribution), or an unpaired nonparametric Mann–Whitney test (non-normal distribution). In addition, in case of testing the difference between COPD GOLDII or GOLDIV patients versus non-COPD patients (three groups), we used a one-way ANOVA and a Dunnett’s post-hoc test for multiple comparisons (normal distribution) or a Kruskal–Wallis test followed by a Dunn’s multiple comparisons test (non-normal distribution). Moreover, the Spearman’s correlation coefficient assessed the correlation between lung function (FEV1/FVC) and transcript levels of PPARGC1B and PPRC1 in human peripheral lung tissue (n = 14) (C, D). Individual subjects are represented by open circles (non-COPD patients), squares (all COPD patients), triangles (COPD GOLDII patients) and diamonds (COPD GOLDIV patients). Statistical significance is indicated as *p < 0.05 compared to non-COPD patients.
Next, we explored the abundance of PPARGC1-coactivated transcription factors in peripheral lung tissue from COPD and non-COPD patients. Although the transcript levels of most transcription factors that we analysed were not different between COPD and non-COPD patients, we observed a significant decrease in mRNA expression of PPAR delta (PPARD) in COPD patients (Fig. 2A,B). This decline in PPARD abundance was in particular observed in peripheral lung tissue from COPD GOLDIV patients relative to non-COPD patients (Fig. 2C). In addition, stratification of the findings according to disease status revealed an increase in the expression of TFAM in COPD GOLDIV patients compared to non-COPD patients (Fig. 2D). We next assessed whether these differences in transcript levels of PPARD and TFAM correlated with lung function parameters. Indeed, correlation analyses revealed a positive correlation between lung function parameters and transcript levels of PPARD (ρ = 0.741, p = 0.002; R2 = 0.344), while a negative correlation was observed between lung function and TFAM expression (ρ = − 0.613, p = 0.020; R2 = 0.432) (Figs. 2E,F). Lastly, we assessed the protein abundance of transcription factors involved in mitochondrial biogenesis and energy metabolism, NRF1 and TFAM, but no significant alterations were observed between COPD and non-COPD patients (Fig. 2G–I) (Figure S1).
Altered expression of PPARGC1-co-activated transcription factors in peripheral lung tissue from very severe COPD patients. Real-time qPCR analysis was used to analyse mRNA expression of PPARA, PPARD and ESRRA (A, C) as well as NRF1, NRF2, and TFAM (B, D) in human peripheral lung tissue from non-COPD patients (n = 5) and COPD patients (n = 9; GOLDII n = 4/9, GOLDIV n = 5/9). Furthermore, Spearman’s correlation coefficient has been determined to evaluate the correlation between lung function (FEV1/FVC) and transcript levels of PPARD or TFAM (E, F) in human peripheral lung tissue (n = 14). Protein levels of NRF1 and TFAM were assessed in human peripheral lung tissue from non-COPD patients (n = 6) and COPD patients (n = 9–10; GOLDII n = 2–3/9–10, GOLDIV n = 7/9–10) by western blot analysis (G-I). For illustration purposes of the protein abundance in the human peripheral lung tissue homogenates, examples of bands of the target proteins along with the corresponding normalization bands of the Ponceau S staining are shown of one patient/group, which is not always representative for the mean and changes in the whole patient group as quantified in the box plots (due to the variation between patients). Black boxes represent a series of bands for specific proteins that were cut from the same blot. Data are presented as mean fold change of COPD patients (GOLDII or GOLDIV) relative to non-COPD patients in box plots (min to max). Statistical differences between COPD patients versus non-COPD patients (two groups) were tested using a two-tailed unpaired parametric t-test (normal distribution) or an unpaired nonparametric Mann–Whitney test (non-normal distribution). In addition, in case of testing the difference between COPD GOLDII or GOLDIV patients versus non-COPD patients (three groups), we used a one-way ANOVA and a Dunnett’s post-hoc test for multiple comparisons (normal distribution) or a Kruskal–Wallis test followed by a Dunn’s multiple comparisons test (non-normal distribution). Individual subjects are represented by open circles (non-COPD patients), squares (all COPD patients), triangles (COPD GOLDII patients) and diamonds (COPD GOLDIV patients). Statistical significance is indicated as *p < 0.05 compared to non-COPD patients.
In summary, this data shows that, with the exception of an upregulation of TFAM mRNA expression, transcript abundance of molecules involved in the regulation of mitochondrial biogenesis by the PPARGC1 family is downregulated in peripheral lung tissue from COPD patients, in particular in COPD GOLDIV patients.
In line with the alterations in peripheral lung tissue as described above, we did observe decreased transcript levels of PPARGC1B between COPD and non-COPD patients in undifferentiated as well as differentiated PBEC cultures. No pronounced changes in the abundance of other molecules controlling mitochondrial biogenesis were observed in both PBEC models (Tables S4 and S5; Figures S2 and S3).
Increased transcript levels of BNIP3L in peripheral lung tissue from COPD patients
Besides mitochondrial biogenesis, mitochondrial quality control by the selective breakdown of damaged or defected mitochondria (i.e., mitophagy) also plays a critical role in maintaining mitochondrial homeostasis. Therefore, we next assessed the protein and mRNA abundance of key molecules involved in both receptor-mediated and ubiquitin-mediated mitophagy in peripheral lung tissue and PBEC from COPD patients.
No pronounced differences in mRNA and protein levels of key regulators associated with ubiquitin-mediated mitophagy were observed in peripheral lung tissue from COPD patients (Fig. 3A–E; Figure S1). Also, there was no correlation between lung function parameters and genes associated with ubiquitin-mediated mitophagy (data not shown). Interestingly, with respect to receptor-mediated mitophagy, significantly increased transcript levels of BNIP3L were observed in peripheral lung tissue from COPD patients, in particular in very severe COPD patients (Figs. 4A,B). This increased expression of BNIP3L negatively correlated with lung function (ρ = − 0.745, p = 0.002; R2 = 0.527) (Fig. 4C). In contrast to transcript levels, no changes were observed in the expression of proteins involved in receptor-mediated mitophagy in lung tissue from COPD patients relative to non-COPD patients (Figs. 4D,F; Figure S1).
The abundance of constituents involved in ubiquitin-mediated mitophagy in peripheral lung tissue from COPD patients. Real-time qPCR analysis was used to analyse mRNA expression of PINK1 and PRKN in human peripheral lung tissue from non-COPD patients (n = 5) and COPD patients (n = 9; GOLDII n = 4/9, GOLDIV n = 5/9) (A, B). Protein levels of PINK1-I, PINK1-II and PRKN were assessed in human peripheral lung tissue from non-COPD patients (n = 6) and COPD patients (n = 10; GOLDII n = 3/10, GOLDIV n = 7/10) by western blot analysis (C-E). Western blot analysis revealed two distinct bands corresponding with expected molecular weights for PINK1 protein, entitled PINK1-I (66 kDa) and PINK1-II (55 kDa). For illustration purposes of the protein abundance in the human peripheral lung tissue homogenates, examples of bands of the target proteins along with the corresponding normalization bands of the Ponceau S staining are shown of one patient/group, which is not always representative for the mean and changes in the whole patient group as quantified in the box plots (due to the variation between patients). The smaller black boxes represent bands of specific proteins from individuals that were cut from the same blot. Data are presented as mean fold change of COPD patients (GOLDII or GOLDIV) relative to non-COPD patients in box plots (min to max). Statistical differences between COPD patients versus non-COPD patients (two groups) were tested using a two-tailed unpaired parametric t-test (normal distribution). In addition, in case of testing the difference between COPD GOLDII or GOLDIV patients versus non-COPD patients (three groups), we used a one-way ANOVA and a Dunnett’s post-hoc test for multiple comparisons (normal distribution) or a Kruskal–Wallis test followed by a Dunn’s multiple comparisons test (non-normal distribution). Individual subjects are represented by open circles (non-COPD patients), squares (all COPD patients), triangles (COPD GOLDII patients) and diamonds (COPD GOLDIV patients). A trend is indicated as #p < 0.1 compared to non-COPD patients.
Increased transcript levels of mitophagy protein BNIP3L in peripheral lung tissue from very severe COPD patients. Real-time qPCR analysis was used to analyse mRNA expression of BNIP3, BNIP3L and FUNDC1 in human peripheral lung tissue from non-COPD patients (n = 5) and COPD patients (n = 9; GOLDII n = 4/9, GOLDIV n = 5/9) (A, B). Moreover, the Spearman’s correlation coefficient has been determined to evaluate the correlation between lung function (FEV1/FVC) and transcript levels of BNIP3L in human peripheral lung tissue (n = 14) (C). Protein levels of BNIP3, BNIP3L, and FUNDC1 were assessed in human peripheral lung tissue from non-COPD patients (n = 6) and COPD patients (n = 10; GOLDII n = 3/10, GOLDIV n = 7/10) by western blot analysis (D–F). For illustration purposes of the protein abundance in the human peripheral lung tissue homogenates, examples of bands of the target proteins along with the corresponding normalization bands of the Ponceau S staining are shown of one patient/group, which is not always representative for the mean and changes in the whole patient group as quantified in the box plots (due to the variation between patients). Black boxes represent a series of bands for specific proteins that were cut from the same blot. Data are presented as mean fold change of COPD patients (GOLDII or GOLDIV) relative to non-COPD patients in box plots (min to max). Statistical differences between COPD patients versus non-COPD patients (two groups) were tested using a two-tailed unpaired parametric t-test (normal distribution), or an unpaired nonparametric Mann–Whitney test (non-normal distribution). In addition, in case of testing the difference between COPD GOLDII or GOLDIV patients versus non-COPD patients (three groups), we used a one-way ANOVA and a Dunnett’s post-hoc test for multiple comparisons (normal distribution). Individual subjects are represented by open circles (non-COPD patients), squares (all COPD patients), triangles (COPD GOLDII patients) and diamonds (COPD GOLDIV patients). Statistical significance is indicated as *p < 0.05 compared to non-COPD patients.
As autophagy is required to facilitate the degradation of mitochondria by formation of an autophagosome, we investigated if the expression of specific autophagy proteins was affected in lung tissue from COPD patients. In summary, we observed no alterations in either protein nor gene expression of molecules involved in autophagy in peripheral lung tissue from COPD patients relative to non-COPD patients (Table 2; Figure S1).
Elevated mRNA levels of BNIP3L, as depicted in Fig. 4, were not observed in undifferentiated or differentiated PBEC from COPD patients. Moreover, in accordance with our other findings in peripheral lung tissue, the abundance of mitophagy and autophagy regulators was not different in PBECs from COPD-patients compared to non-COPD patients (Tables S4 and S5; Figs. S2 and S3).
With regard to mitophagy and autophagy, in summary, the expression of BNIP3L is elevated in peripheral lung tissue, not in PBEC, from very severe COPD patients , while other markers associated with mitophagy and autophagy were not differently-expressed in COPD patients relative to non-COPD patients.
No pronounced changes in the expression of constituents of mitochondrial metabolic processes and mitochondrial dynamics in peripheral tissue or PBEC from COPD patients
Given the changes in expression of markers of mitochondrial biogenesis and mitophagy, we investigated transcript and protein abundance of a myriad of constituents of different mitochondrial metabolic processes including the electron transport chain, tricarboxylic acid cycle, glycolysis, and fatty acid β-oxidation. As shown in Table 3 and Figure S1, no pronounced differences in abundance of these regulators were observed in peripheral lung tissue from COPD patients versus non-COPD patients, except for minor changes in the transcript abundance of subunits of complex III (decrease) and complex IV (increase). Lastly, we explored the regulation of mitochondrial dynamics and observed unaltered expression of fission- and fusion-associated proteins in peripheral lung tissue from COPD patients, sustained during different disease stages (Table 4; Figure S1). In line with these results, we did not observe pronounced alterations in the regulation of mitochondrial content, metabolism or dynamics in undifferentiated or differentiated PBEC cultures from COPD-patients compared to non-COPD patients, except a downregulation of OPA1 expression (mitochondrial fission) in differentiated PBEC from COPD patients (Tables S4 and S5; Figs. S2 and S3).
Discussion
Abnormalities in mitochondrial morphology and content have been described in PBECs from COPD patients. However, whether or not the molecular pathways controlling this (mitochondrial biogenesis versus mitophagy) are altered, thus far was unknown. Moreover, if these changes also occur in peripheral lung tissue is unexplored. Therefore, in this study, as our primary aim, we evaluated for the first time, the protein and transcript abundance of a broad spectrum of molecules involved in the regulation of mitochondrial biogenesis, mitophagy, mitochondrial dynamics and mitochondrial metabolism in both peripheral lung tissue from COPD patients at different stages of the disease and in undifferentiated and differentiated PBEC from COPD patients and non-COPD patients. As our main findings, we observed a significant reduction in transcript levels of molecules controlling mitochondrial biogenesis and an upregulation of a molecule involved in receptor-mediated mitophagy (BNIP3L), in peripheral lung tissue from very severe COPD patients. In general, these changes were not recapitulated in PBEC from COPD patients, with the exception of decreased PPARGC1B expression which was observed in both PBEC models. An overview of all data is presented in Fig. 5.
Our key observation was a significant decrease in transcript levels of molecules essentially involved in mitochondrial biogenesis in peripheral lung tissue and PBECs from COPD patients, especially in a very severe disease stage. Although the investigation of a comprehensive set of regulatory molecules involved in mitochondrial biogenesis has not been previously described, specific subsets of regulatory molecules have been investigated in PBEC or lung homogenates from COPD patients previously. In line with our data, progressively decreased expression of PPARGC1A was shown in lung homogenates of moderate and severe COPD patients (GOLDII and GOLDIII), while increased expression levels were observed in mild COPD patients (GOLDI) compared to control subjects38. In contrast, increased transcript levels of PPARGC1A have been observed in PBEC of ex-smoking COPD GOLDIV patients relative to never-smoking or smoking controls19. Regarding the impact of cigarette smoke on the regulation of mitochondrial biogenesis, higher mRNA levels of PPARGC1A and PPRC1 and lower mRNA levels of PPARGC1B have been reported differentiated PBECs upon acute whole CS-exposure39. Similar findings were reported in in bronchial epithelial cells s exposed to CS extract40. In addition, decreased expression of PPARGC1A has been observed in experimental models of CS-induced COPD, e.g., mice instilled with CS extract41 or in mice instilled with lipopolysaccharide and CS42, while elevated abundance of PPARGC1A have been observed in rats treated with CS+ lipopolysaccharide38.
In addition, in our study, a key molecule involved in transcription of the mitochondrial genome, TFAM, was found to be increased in peripheral lung tissue from very severe COPD patients compared to non-COPD patients. These differences were previously not observed in lung homogenates from ex-smoking severe COPD GOLDIV patients relative to never-smoking controls19. Moreover, in contrast to our findings, decreased TFAM levels were previously observed in lung tissue from COPD patients compared to non-COPD patients43. These apparently contradictory findings i could be partly explained by differences in disease status of these study populations. Also, as we previously reported that CS can increase Tfam mRNA expression39, an impact of smoke exposure on the abundance of these transcription factors cannot be excluded. In this context, increased expression levels may be evidence of a compensatory cellular response to induce mitochondrial biogenesis.
Although the directionality of these alterations may depend on the cell type, disease status or time or dose of exposure, this collective body of evidence demonstrates that the molecular regulation of mitochondrial biogenesis is disturbed in COPD patients and is impacted by smoking in epithelial cells of both the bronchial and alveolar compartment. The fact that some of our read-out parameters correlated (inversely) with lung function parameters suggests that the regulation of mitochondrial metabolism/turnover may be important for maintaining adequate lung function or that impairments in (the regulation of) mitochondrial content/function may be implicated in deterioration of lung function in COPD. These notions have been suggested in literature previously18,44,45.
For ubiquitin-mediated mitophagy, we found no significant alterations in the abundance of any of the molecules involved in these pathways. In contrast, it has been previously described that PINK1 expression was elevated in lung tissue homogenates from COPD patients19,46,47. In addition, a downregulation of PRKN protein was observed in lung homogenates from COPD patients compared to non-COPD smokers, along with a positive correlation between PRKN protein levels and percentage FEV1/FVC46. These alterations would suggest aberrant ubiquitin-mediated mitophagy in COPD. Moreover, decreased PRKN protein levels were shown smokers relative to non-smokers as well as in mitochondria isolated from mice lung exposed to CS for 6 months indicating impaired PRKN translocation to mitochondria48. Increased PINK1 and decreased PRKN expression was also observed in PBEC cultures acutely exposed to CS39. These data imply a role for smoke exposure in changes in ubiquitin-mediated mitophagy in COPD.. The reason for a lack of alterations in the abundance of these molecules in our study is unclear but disease severity or smoking history may play a role. In addition, the relevance and exact implication of potential changes in this mitophagy pathway in COPD pathogenesis remains to be established, however it is likely that ubiquitin-mediated mitophagy may play a role as it has been shown that animals deficient for PRKN display an increased susceptibility for developing CS-induced emphysema49.
Next to ubiquitin-mediated mitophagy, we also explored other known mitophagy pathways. In this regard, we observed increased mRNA levels of receptor-mediated mitophagy-associated protein BNIP3L in peripheral lung tissue from very severe COPD patients. To our knowledge, this is the first time this has been investigated in airway tissues/cells from COPD patients. Interestingly, we recently showed that the abundance of proteins and genes associated with receptor-mediated mitophagy were increased in PBECs following acute exposure to CS39 which is in line with several other in vitro and in vivo studies50,51. CS-induced hypoxia may play a role in mediating this as previous studies showed that CS stimulates hypoxia-related signalling in airway cell models52,53 which is a known activating pathway for receptor-mediated mitophagy/autophagy54,55,56,57,58,59.
Interestingly, we observed altered gene expression profiles suggestive of impaired ciliogenesis and increased goblet cell formation in COPD PBEC cultures compared to non-COPD cultures. Alterations in the composition of these cell populations have been previously observed in the airway epithelium of COPD patients and smokers and in vitro in small airway epithelial cells exposed to CS. Changes observed herein included an increased number of basal and secretory cells, impaired length/number of ciliated cells, and metaplasia of squamous cells7,8,9,10,60. Both smoking and the disease itself has been suggested to be a contributing factor to these changes in COPD patients7.
No differences were observed in TEER development after differentiation of PBECs between COPD and non-COPD patients. It should be noted however that TEER is just an indication of barrier integrity and not for cellular composition or functionality of the epithelial layer. Some studies showed dysfunctional cilia in PBECs from COPD patients11,12 and reduced beating frequency was shown in nasal cilia from COPD patients13. Also, a significant association has been found between mucus hypersecretion and lung function decline in COPD patients14. The exact pathways underlying this are unknown, but there may be a role for mitochondrial dysfunction herein. We performed correlation analyses to explore correlations between the abundance of mitochondrial markers and markers associated with muco-ciliary function in differentiated PBEC from COPD versus non-COPD patients, but failed to observe convincing correlations. Further research into the relationship between mucociliary function, mucus secretion, and the role of mitochondrial dysfunction herein is required in COPD patients.
Obviously, our study has limitations. Firstly, obtaining and analyzing both undifferentiated- and differentiated PBEC cultures as well as peripheral lung tissue from the same (non)COPD patients would have been ideal. Unfortunately, we were limited to retrieving our biological material from 3 different patient cohorts, which likely introduced variability in our results. Secondly, it must be noted that the peripheral lung tissue as well as non-COPD PBECs used for the undifferentiated PBEC model were isolated from subjects undergoing lung cancer resection surgery. Although material was collected from locations as distant as possible from the tumor, obviously these subjects were not healthy which may have influenced our results. Thirdly, changes in the regulatory pathways that we describe are not necessarily reflective of actual mitochondrial function. Lastly, our 2 of our 3 models or biological material consisted of multiple different cell types, obscuring cell-type specific responses61,62. Future studies using microdissection of the lung tissue or singe-cell sequencing, as well as (co-)cultures of several cell populations may contribute to the understanding of the regulation of mitochondrial turnover and metabolism and the involvement of specific cell types in the context of COPD.
Another observation from our study is the discrepancy in transcript versus protein expression in peripheral lung tissue from non-COPD patients and COPD patients. Whether these discrepancies can be explained by (smoking-induced) changes in post-transcriptional63, remains to be established. Additional explanations could be the low number of patients included in the study and the biological variation. With regard to discrepancies in mRNA and protein levels of mitophagy constituents, the fact that mitophagy proteins are actively broken down together with the mitochondrion during mitophagy and need to be replenished (by transcriptional) upregulation of the respective genes, may contribute to apparently discrepant findings. This also warrants caution in the interpretation of these findings with regard to activation status of the mitophagy process.
In addition to these considerations, some differences in gender and age were identified between moderate and very severe COPD patients that we included in our study. The observation of a more pronounced downregulation in the expression of regulators controlling mitochondrial biogenesis in very severe COPD patients (female; younger age) versus moderate COPD patients (male; older age), is in line with a recent study reporting that genes related to mitochondrial biogenesis/function were differently expressed in younger and older smokers, and in COPD patients compared to non-smokers64. Interestingly, recent evidence also supports a role for accelerated ageing and related mitochondrial dysfunction in the pathogenesis of COPD17. Furthermore, other studies also point to a significant impact of gender on mitochondrial pathways22,65,66.
As a final point, it must be emphasized that our observations are descriptive in nature and do not imply causal involvement of changes in these pathways in COPD pathogenesis. As a decrease in PPARGC1B was the most consistent finding in our study, we attempted to address any causal implication by knocking down of this protein in non-COPD PBECs but unfortunately failed to achieve sufficient knock-down. Difficulties knock-down in PBECs have been described previously67. However, based on literature it can be speculated that, given the well-known role of PPARGC1B (and its better known family member PPARGC1A) in control of mitochondrial function/content, loss of this co-activator molecule likely leads to loss of mitochondrial functionality and content26,27,28. In addition, although mitochondrial abnormalities have been described in PBECs and now also in peripheral lung tissue of COPD patients, whether or not these are involved in COPD pathogenesis is unclear. In this context however, it is interesting to note that mitochondrial dysfunction is well-known to lead to oxidative stress, inflammation, cell death and senescence, all of which are hallmark features in cells of the airways and lungs in COPD. Also, the role of mitochondrial dysfunction in the pathogenesis of lung disease, including COPD, has been extensively reviewed previously18,19. Moreover, using a mouse model of COPD (smoking mice) Cloonan et al. very elegantly showed that preventing cigarette smoke-induced mitochondrial dysfunction ameliorated COPD features in these animals which does suggest causal involvement of mitochondrial dysfunction in COPD pathogenesis44.
In conclusion, our study shows changes in the expression of genes controlling mitochondrial biogenesis and receptor-mediated mitophagy in peripheral lung tissue from very severe COPD patients, along with a correlation with lung function. In general, these results were not observed in undifferentiated and differentiated PBEC from COPD patients, except decreased PPARGC1B transcript levels in both PBEC models. If these changes are involved in the airway pathophysiology of COPD, contribute to aberrant mitochondrial function or may represent compensatory mechanisms remains to be established.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ACTB:
-
Actin B
- AEGM:
-
Airway epithelial cell growth medium
- ALI:
-
Air–liquid interface
- ATP5F1A:
-
ATP synthase F1 subunit alpha
- B2M:
-
Beta-2 microglobulin,
- BEGM:
-
Bronchial epithelial cell growth medium
- BSA:
-
Bovine serum albumin
- BMI:
-
Body mass index
- BNIP3:
-
BCL2 interacting protein 3
- BNIP3L:
-
BCL2 Interacting protein 3-Like
- COPD:
-
Chronic obstructive pulmonary disease
- CS:
-
Cigarette smoke
- COXII:
-
Cytochrome C oxidase subunit II
- COXIII:
-
Cytochrome C oxidase subunit III
- COXIV:
-
Cytochrome C oxidase subunit 4I1
- CYC1:
-
Cytochrome C-1
- CYTB:
-
Mitochondrially encoded cytochrome B
- DNM1L:
-
Dynamin-1-Like
- ESRRA:
-
Estrogen related receptor, alpha
- FEV1:
-
Forced expiratory volume in first second
- FIS1:
-
Fission, mitochondrial 1
- FOXJ1:
-
Forkhead box J1
- FUNDC1:
-
FUN14 domain containing 1
- FVC:
-
Forced vital capacity
- GABARAPL1:
-
GABA type a receptor associated protein like 1
- HADHA:
-
Hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha
- HBSS:
-
Hank’s balanced salt solution
- HK2:
-
Hexokinase 2
- KRT5:
-
Keratin 5
- KRT14:
-
Keratin 14
- MAP1LC3A:
-
Microtubule-associated protein 1 light chain 3 alpha
- MAP1LC3B:
-
Microtubule-associated protein 1 light chain 3 beta
- MFN1:
-
Mitofusin 1
- MFN2:
-
Mitofusin 2
- MUC5AC:
-
Mucin 5AC, oligomeric mucus/gel-forming
- NDUFB3:
-
NADH:Ubiquinone oxidoreductase subunit B3
- NDUFB8:
-
NADH:Ubiquinone oxidoreductase subunit B8
- NRF1:
-
Nuclear respiratory factor 1,
- NRF2:
-
Nuclear respiratory factor 2
- OPA1:
-
OPA1, Mitochondrial dynamin like GTPase
- OXPHOS:
-
Oxidative phosphorylation
- PBEC:
-
Primary bronchial epithelial cells
- PDK4:
-
Pyruvate dehydrogenase kinase 4
- PINK1:
-
PTEN Induced kinase 1
- PPAR:
-
Peroxisome proliferator-activated receptor
- PPARA:
-
Peroxisome proliferator activated receptor alpha
- PPARD:
-
Peroxisome proliferator activated receptor delta
- PPARG:
-
Peroxisome proliferator-activated receptor gamma
- PPARGC1:
-
Peroxisome proliferator-activated receptor gamma, coactivator 1
- PPARGC1A:
-
Peroxisome proliferator-activated receptor gamma, coactivator 1 Alpha
- PPARGC1B:
-
Peroxisome proliferator-activated receptor gamma, coactivator 1 beta
- PPIA:
-
Peptidylprolyl isomerase A
- PPRC1:
-
Peroxisome proliferator-activated receptor gamma, coactivator-related 1
- PRKN:
-
Parkin RBR E3 ubiquitin protein ligase
- RPL13A:
-
Ribosomal protein L13A
- SCGB1A1:
-
Secretoglobin family 1a member 1
- SDHB:
-
Succinate dehydrogenase complex iron sulfur subunit B
- SEM:
-
Standard error of the mean
- SQSTM1:
-
Sequestosome 1, transcript variant 1/4
- TBST:
-
Tween20 Tris-buffered saline
- TEER:
-
Transepithelial electrical resistance
- TFAM:
-
Transcription Factor A, mitochondrial
- TOMM20:
-
Translocase of outer mitochondrial membrane 20
- TP63:
-
Tumor protein P63
- UQCRC2:
-
Ubiquinol-cytochrome C reductase core protein 2
References
GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the global burden of disease study 2015. Lancet 388, 1459–1544. https://doi.org/10.1016/s0140-6736(16)31012-1 (2016).
GBD 2019 Diseases and Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the global burden of disease study 2019. Lancet 396, 1204–1222. https://doi.org/10.1016/s0140-6736(20)30925-9 (2020).
Vestbo, J. et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 187, 347–365. https://doi.org/10.1164/rccm.201204-0596PP (2013).
Hogg, J. C. Pathophysiology of airflow limitation in chronic obstructive pulmonary disease. Lancet 364, 709–721. https://doi.org/10.1016/s0140-6736(04)16900-6 (2004).
Global Initiative For Chronic Obstructive Lung Disease. Global Strategy For Diagnosis, Management, And Prevention Of Chronic Obstructive Lung Disease: Report v1.0 (2022).
Hiemstra, P. S., Grootaers, G., van der Does, A. M., Krul, C. A. M. & Kooter, I. M. Human lung epithelial cell cultures for analysis of inhaled toxicants: Lessons learned and future directions. Toxicol. In Vitro 47, 137–146. https://doi.org/10.1016/j.tiv.2017.11.005 (2018).
Rigden, H. M. et al. Squamous metaplasia is increased in the bronchial epithelium of smokers with chronic obstructive pulmonary disease. PLoS ONE 11, e0156009–e0156009. https://doi.org/10.1371/journal.pone.0156009 (2016).
Crystal, R. G. Airway basal cells. The smoking gun of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 190, 1355–1362 (2014).
Saetta, M. T. G. et al. Goblet cell hyperplasia and epithelial inflammation in peripheral airways of smokers with both symptoms of chronic bronchitis and chronic airflow limitation. Am. J. Respire. Crit. Care Med. 161, 1016–1021 (2000).
Jeffery, P. K. Comparison of the structural and inflammatory features of COPD and asthma Giles F Filley Lecture. Chest 117, 251S-260S (2000).
Ancel, J. et al. Impaired ciliary beat frequency and ciliogenesis alteration during airway epithelial cell differentiation in COPD. Diagnostics (Basel) https://doi.org/10.3390/diagnostics11091579 (2021).
Thomas, B. et al. Dysfunctional bronchial cilia are a feature of chronic obstructive pulmonary disease (COPD). Copd 18, 657–663. https://doi.org/10.1080/15412555.2021.1963695 (2021).
Yaghi, A., Zaman, A., Cox, G. & Dolovich, M. B. Ciliary beating is depressed in nasal cilia from chronic obstructive pulmonary disease subjects. Respir. Med. 106, 1139–1147. https://doi.org/10.1016/j.rmed.2012.04.001 (2012).
Vestbo, J., Prescott, E., Lange, P., Copenhagen City Heart Study Group. Association of chronic mucus hypersecretion with FEV1 decline and chronic obstructive pulmonary disease morbidity. Am. J. Respir. Crit. Care Med. 153, 1530–1535. https://doi.org/10.1164/ajrccm.153.5.8630597 (1996).
Higham, A., Quinn, A. M., Cançado, J. E. D. & Singh, D. The pathology of small airways disease in COPD: Historical aspects and future directions. Respir. Res. 20, 49. https://doi.org/10.1186/s12931-019-1017-y (2019).
MacNee, W. Pathogenesis of chronic obstructive pulmonary disease. Clin. Chest Med. 28, 479–513. https://doi.org/10.1016/j.ccm.2007.06.008 (2007).
Cloonan, S. M., Kim, K., Esteves, P., Trian, T. & Barnes, P. J. Mitochondrial dysfunction in lung ageing and disease. Eur. Respir. Rev. https://doi.org/10.1183/16000617.0165-2020 (2020).
Cloonan, S. M. & Choi, A. M. Mitochondria in lung disease. J. Clin. Invest. 126, 809–820. https://doi.org/10.1172/jci81113 (2016).
Hoffmann, R. F. et al. Prolonged cigarette smoke exposure alters mitochondrial structure and function in airway epithelial cells. Respir. Res. 14, 97. https://doi.org/10.1186/1465-9921-14-97 (2013).
Hara, H. et al. Mitochondrial fragmentation in cigarette smoke-induced bronchial epithelial cell senescence. Am. J. Physiol. Lung Cell. Mol. Physiol. 305, L737-746. https://doi.org/10.1152/ajplung.00146.2013 (2013).
Zhang, W. Z. et al. Association of plasma mitochondrial DNA with COPD severity and progression in the SPIROMICS cohort. Respir. Res. 22, 126. https://doi.org/10.1186/s12931-021-01707-x (2021).
Zhang, W. Z. et al. Association of urine mitochondrial DNA with clinical measures of COPD in the SPIROMICS cohort. JCI Insight 5, e133984. https://doi.org/10.1172/jci.insight.133984 (2020).
Sundar, I. K., Maremanda, K. P. & Rahman, I. Mitochondrial dysfunction is associated with Miro1 reduction in lung epithelial cells by cigarette smoke. Toxicol. Lett. 317, 92–101. https://doi.org/10.1016/j.toxlet.2019.09.022 (2019).
Ballweg, K., Mutze, K., Königshoff, M., Eickelberg, O. & Meiners, S. Cigarette smoke extract affects mitochondrial function in alveolar epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 307, L895-907. https://doi.org/10.1152/ajplung.00180.2014 (2014).
Ploumi, C., Daskalaki, I. & Tavernarakis, N. Mitochondrial biogenesis and clearance: A balancing act. Febs J. 284, 183–195. https://doi.org/10.1111/febs.13820 (2017).
Jornayvaz, F. R. & Shulman, G. I. Regulation of mitochondrial biogenesis. Essays Biochem. 47, 69–84. https://doi.org/10.1042/bse0470069 (2010).
Lin, J., Handschin, C. & Spiegelman, B. M. Metabolic control through the PGC-1 family of transcription coactivators. Cell. Metab. 1, 361–370. https://doi.org/10.1016/j.cmet.2005.05.004 (2005).
Scarpulla, R. C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta https://doi.org/10.1016/j.bbamcr.2010.09.019 (1813).
Gomes, L. C. & Scorrano, L. Mitochondrial morphology in mitophagy and macroautophagy. Biochim. Biophys. Acta 205–212, 2013. https://doi.org/10.1016/j.bbamcr.2012.02.012 (1833).
Fritsch, L. E., Moore, M. E., Sarraf, S. A. & Pickrell, A. M. Ubiquitin and receptor-dependent mitophagy pathways and their implication in neurodegeneration. J. Mol. Biol. 432, 2510–2524. https://doi.org/10.1016/j.jmb.2019.10.015 (2020).
Detmer, S. A. & Chan, D. C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8, 870–879. https://doi.org/10.1038/nrm2275 (2007).
Ryter, S. W. et al. Mitochondrial dysfunction as a pathogenic mediator of chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Ann. Am. Thorac. Soc. 15, S266-s272. https://doi.org/10.1513/AnnalsATS.201808-585MG (2018).
Hara, H., Kuwano, K. & Araya, J. Mitochondrial quality control in COPD and IPF. Cells https://doi.org/10.3390/cells7080086 (2018).
Aghapour, M. et al. Mitochondria: At the crossroads of regulating lung epithelial cell function in chronic obstructive pulmonary disease. Am. J. Physiol. Lung Cell. Mol. Physiol. 318, L149-l164. https://doi.org/10.1152/ajplung.00329.2019 (2020).
van Wetering, S. et al. Regulation of SLPI and elafin release from bronchial epithelial cells by neutrophil defensins. Am. J. Physiol. Lung Cell. Mol. Physiol. 278, L51-58. https://doi.org/10.1152/ajplung.2000.278.1.L51 (2000).
van Wetering, S. et al. Epithelial differentiation is a determinant in the production of eotaxin-2 and -3 by bronchial epithelial cells in response to IL-4 and IL-13. Mol. Immunol. 44, 803–811. https://doi.org/10.1016/j.molimm.2006.04.008 (2007).
Karwelat, D. et al. Influenza virus-mediated suppression of bronchial chitinase-3-like 1 secretion promotes secondary pneumococcal infection. Faseb J. 34, 16432–16448. https://doi.org/10.1096/fj.201902988RR (2020).
Li, J., Dai, A., Hu, R., Zhu, L. & Tan, S. Positive correlation between PPARgamma/PGC-1alpha and gamma-GCS in lungs of rats and patients with chronic obstructive pulmonary disease. Acta Biochim. Biophys. Sin. 42, 603–614. https://doi.org/10.1093/abbs/gmq071 (2010).
Tulen, C. B. M. et al. Dysregulated mitochondrial metabolism upon cigarette smoke exposure in various human bronchial epithelial cell models. Dis. Model Mech. https://doi.org/10.1242/dmm.049247 (2022).
Vanella, L. et al. A new antioxidant formulation reduces the apoptotic and damaging effect of cigarette smoke extract on human bronchial epithelial cells. Eur. Rev. Med. Pharmacol. Sci. 21, 5478–5484. https://doi.org/10.26355/eurrev_201712_13938 (2017).
Wang, S. et al. Function of hesperidin alleviating inflammation and oxidative stress responses in COPD mice might be related to SIRT1/PGC-1alpha/NF-kappaB signaling axis. J. Recept. Signal Transduct. Res. https://doi.org/10.1080/10799893.2020.1738483 (2020).
Wang, X. L., Li, T., Li, J. H., Miao, S. Y. & Xiao, X. Z. The effects of resveratrol on inflammation and oxidative stress in a rat model of chronic obstructive pulmonary disease. Molecules https://doi.org/10.3390/molecules22091529 (2017).
Peng, H. et al. Expression and methylation of mitochondrial transcription factor a in chronic obstructive pulmonary disease patients with lung cancer. PLoS One 8, e82739. https://doi.org/10.1371/journal.pone.0082739 (2013).
Cloonan, S. M. et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat. Med. 22, 163–174. https://doi.org/10.1038/nm.4021 (2016).
Yang, D. et al. Mitoquinone ameliorates cigarette smoke-induced airway inflammation and mucus hypersecretion in mice. Int. Immunopharmacol. 90, 107149. https://doi.org/10.1016/j.intimp.2020.107149 (2021).
Ito, S. et al. PARK2-mediated mitophagy is involved in regulation of HBEC senescence in COPD pathogenesis. Autophagy 11, 547–559. https://doi.org/10.1080/15548627.2015.1017190 (2015).
Mizumura, K. et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J. Clin. Invest. 124, 3987–4003. https://doi.org/10.1172/jci74985 (2014).
Ahmad, T. et al. Impaired mitophagy leads to cigarette smoke stress-induced cellular senescence: Implications for chronic obstructive pulmonary disease. FASEB J. 29, 2912–2929. https://doi.org/10.1096/fj.14-268276 (2015).
Araya, J. et al. PRKN-regulated mitophagy and cellular senescence during COPD pathogenesis. Autophagy 15, 510–526. https://doi.org/10.1080/15548627.2018.1532259 (2019).
Wen, W. et al. Silencing FUNDC1 alleviates chronic obstructive pulmonary disease by inhibiting mitochondrial autophagy and bronchial epithelium cell apoptosis under hypoxic environment. J. Cell. Biochem. 120, 17602–17615. https://doi.org/10.1002/jcb.29028 (2019).
Zhang, M. et al. Nix/BNIP3L-dependent mitophagy accounts for airway epithelial cell injury induced by cigarette smoke. J. Cell. Physiol. 234, 14210–14220. https://doi.org/10.1002/jcp.28117 (2019).
Daijo, H. et al. Cigarette smoke reversibly activates hypoxia-inducible factor 1 in a reactive oxygen species-dependent manner. Sci. Rep. 6, 34424–34424. https://doi.org/10.1038/srep34424 (2016).
Zhang, Q. et al. Nicotine induces hypoxia-inducible factor-1α expression in human lung cancer cells via nicotinic acetylcholine receptor–mediated signaling pathways. Clin. Cancer Res. 13, 4686–4694 (2007).
Bellot, G. et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell. Biol. 29, 2570–2581. https://doi.org/10.1128/mcb.00166-09 (2009).
Ishihara, M. et al. Sestrin-2 and BNIP3 regulate autophagy and mitophagy in renal tubular cells in acute kidney injury. Am. J. Physiol. Renal Physiol. 305, F495-509. https://doi.org/10.1152/ajprenal.00642.2012 (2013).
Lee, S.-J., Kim, H.-P., Jin, Y., Choi, A. M. K. & Ryter, S. W. Beclin 1 deficiency is associated with increased hypoxia-induced angiogenesis. Autophagy 7, 829–839. https://doi.org/10.4161/auto.7.8.15598 (2011).
Lv, M. et al. Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy. Protein Cell 8, 25–38. https://doi.org/10.1007/s13238-016-0328-8 (2017).
Wang, L. et al. Ulk1/FUNDC1 prevents nerve cells from hypoxia-induced apoptosis by promoting cell autophagy. Neurochem. Res. 43, 1539–1548. https://doi.org/10.1007/s11064-018-2568-x (2018).
Wu, W. et al. FUNDC1 is a novel mitochondrial-associated-membrane (MAM) protein required for hypoxia-induced mitochondrial fission and mitophagy. Autophagy 12, 1675–1676. https://doi.org/10.1080/15548627.2016.1193656 (2016).
Gindele, J. A. et al. Intermittent exposure to whole cigarette smoke alters the differentiation of primary small airway epithelial cells in the air-liquid interface culture. Sci. Rep. 10, 6257. https://doi.org/10.1038/s41598-020-63345-5 (2020).
Khan, Y. & Lynch, D. Histology, Lung (StatPearls Publishing, 2022).
Altorki, N. K. et al. The lung microenvironment: An important regulator of tumour growth and metastasis. Nat. Rev. Cancer 19, 9–31. https://doi.org/10.1038/s41568-018-0081-9 (2019).
Aloufi, N., Alluli, A., Eidelman, D. H. & Baglole, C. J. Aberrant post-transcriptional regulation of protein expression in the development of chronic obstructive pulmonary disease. Int. J. Mol. Sci. https://doi.org/10.3390/ijms222111963 (2021).
Maremanda, K. P., Sundar, I. K., Li, D. & Rahman, I. Age-dependent assessment of genes involved in cellular senescence, telomere, and mitochondrial pathways in human lung tissue of smokers, COPD, and IPF: Associations with SARS-CoV-2 COVID-19 ACE2-TMPRSS2-Furin-DPP4 Axis. Front. Pharmacol. 11, 584637. https://doi.org/10.3389/fphar.2020.584637 (2020).
Koo, H. K. et al. Sex-specific associations with DNA methylation in lung tissue demonstrate smoking interactions. Epigenetics 16, 692–703. https://doi.org/10.1080/15592294.2020.1819662 (2021).
Glass, K. et al. Sexually-dimorphic targeting of functionally-related genes in COPD. BMC Syst. Biol. 8, 1–17 (2014).
Platz, J. et al. Application of small interfering RNA (siRNA) for modulation of airway epithelial gene expression. Oligonucleotides 15, 132–138. https://doi.org/10.1089/oli.2005.15.132 (2015).
Acknowledgements
The authors would like to thank Dr. Mieke A. Dentener and Nico Kloosterboer of the Primary Lung Culture Facility (PLUC; Maastricht University Medical Center +) as well as Dr. Evelyn Vollmeister and Isabell Beinborn from the Institute for Lung Research (iLung; Philipps-University Marburg) for providing donor cells, advice, and protocols.
Funding
This research is supported by the Netherlands Food and Consumer Product Safety Authority (NVWA).
Author information
Authors and Affiliations
Contributions
Conceptualization: Tulen, C.B.M., van de Wetering, C., Schiffers, C.H.J., Benedikter, B.J., Schmeck, B.T., Reynaert, N.L., Opperhuizen, A, van Schooten, F.J., Remels, A.H.V. Investigation and formal analysis: Tulen, C.B.M., van de Wetering, C., Schiffers, C.H.J., Benedikter, B.J., Leermakers, P.A., Boukhaled, J.H., Drittij, M.J, Reynaert, N.L. Writing – Drafting: Tulen, C.B.M. Writing – Revising: van de Wetering, C., Schiffers, C.H.J., Benedikter, B.J., Leermakers, P.A., Boukhaled, J.H., Drittij, M.J., Schmeck, B.T., Reynaert, N.L., Opperhuizen, A, van Schooten, F.J., Remels, A.H.V, Weltjens E. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tulen, C.B.M., van de Wetering, C., Schiffers, C.H.J. et al. Alterations in the molecular control of mitochondrial turnover in COPD lung and airway epithelial cells. Sci Rep 14, 4821 (2024). https://doi.org/10.1038/s41598-024-55335-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-55335-8
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.